HERV-K(HML7) Integrations in the Human Genome: Comprehensive Characterization and Comparative Analysis in Non-Human Primates
- PMID: 34069102
- PMCID: PMC8156875
- DOI: 10.3390/biology10050439
HERV-K(HML7) Integrations in the Human Genome: Comprehensive Characterization and Comparative Analysis in Non-Human Primates
Abstract
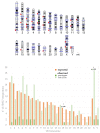
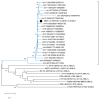
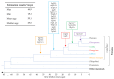
Endogenous Retroviruses (ERVs) are ancient relics of infections that affected the primate germ line and constitute about 8% of our genome. Growing evidence indicates that ERVs had a major role in vertebrate evolution, being occasionally domesticated by the host physiology. In addition, human ERV (HERV) expression is highly investigated for a possible pathological role, even if no clear associations have been reported yet. In fact, on the one side, the study of HERV expression in high-throughput data is a powerful and promising tool to assess their actual dysregulation in diseased conditions; but, on the other side, the poor knowledge about the various HERV group genomic diversity and individual members somehow prevented the association between specific HERV loci and a given molecular mechanism of pathogenesis. The present study is focused on the HERV-K(HML7) group that-differently from the other HERV-K members-still remains poorly characterized. Starting from an initial identification performed with the software RetroTector, we collected 23 HML7 proviral insertions and about 160 HML7 solitary LTRs that were analyzed in terms of genomic distribution, revealing a significant enrichment in chromosome X and the frequent localization within human gene introns as well as in pericentromeric and centromeric regions. Phylogenetic analyses showed that HML7 members form a monophyletic group, which based on age estimation and comparative localization in non-human primates had its major diffusion between 20 and 30 million years ago. Structural characterization revealed that besides 3 complete HML7 proviruses, the other group members shared a highly defective structure that, however, still presents recognizable functional domains, making it worth further investigation in the human population to assess the presence of residual coding potential.
Keywords: HERV; HERV-K; HML7; endogenous retroviruses; retrotransposons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification of a novel HERV-K(HML10): comprehensive characterization and comparative analysis in non-human primates provide insights about HML10 proviruses structure and diffusion.Mob DNA. 2017 Nov 2;8:15. doi: 10.1186/s13100-017-0099-7. eCollection 2017. Mob DNA. 2017. PMID: 29118853 Free PMC article.
-
Identification, comprehensive characterization, and comparative genomics of the HERV-K(HML8) integrations in the human genome.Virus Res. 2023 Jan 2;323:198976. doi: 10.1016/j.virusres.2022.198976. Epub 2022 Oct 26. Virus Res. 2023. PMID: 36309315 Free PMC article.
-
Phylogenetic Analysis Reveals That ERVs "Die Young" but HERV-H Is Unusually Conserved.PLoS Comput Biol. 2016 Jun 13;12(6):e1004964. doi: 10.1371/journal.pcbi.1004964. eCollection 2016 Jun. PLoS Comput Biol. 2016. PMID: 27295277 Free PMC article.
-
Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses.Front Immunol. 2018 Sep 10;9:2039. doi: 10.3389/fimmu.2018.02039. eCollection 2018. Front Immunol. 2018. PMID: 30250470 Free PMC article. Review.
-
Type W Human Endogenous Retrovirus (HERV-W) Integrations and Their Mobilization by L1 Machinery: Contribution to the Human Transcriptome and Impact on the Host Physiopathology.Viruses. 2017 Jun 27;9(7):162. doi: 10.3390/v9070162. Viruses. 2017. PMID: 28653997 Free PMC article. Review.
Cited by
-
Long-term host-pathogen evolution of endogenous beta- and gammaretroviruses in mouse lemurs with little evidence of recent retroviral introgression.Virus Evol. 2022 Dec 14;9(1):veac117. doi: 10.1093/ve/veac117. eCollection 2023. Virus Evol. 2022. PMID: 36632481 Free PMC article.
-
An assessment of bioinformatics tools for the detection of human endogenous retroviral insertions in short-read genome sequencing data.Front Bioinform. 2023 Feb 8;2:1062328. doi: 10.3389/fbinf.2022.1062328. eCollection 2022. Front Bioinform. 2023. PMID: 36845320 Free PMC article.
-
Comprehensive Identification and Characterization of HML-9 Group in Chimpanzee Genome.Viruses. 2024 May 31;16(6):892. doi: 10.3390/v16060892. Viruses. 2024. PMID: 38932184 Free PMC article.
-
A Systems Biology Approach on the Regulatory Footprint of Human Endogenous Retroviruses (HERVs).Diseases. 2022 Nov 2;10(4):98. doi: 10.3390/diseases10040098. Diseases. 2022. PMID: 36412592 Free PMC article.
-
Human endogenous retroviruses in development and disease.Comput Struct Biotechnol J. 2021 Nov 2;19:5978-5986. doi: 10.1016/j.csbj.2021.10.037. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34849202 Free PMC article. Review.
References
-
- Blond J.-L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., Cosset F.-L. An Envelope Glycoprotein of the Human Endogenous Retrovirus HERV-W Is Expressed in the Human Placenta and Fuses Cells Expressing the Type D Mammalian Retrovirus Receptor. J. Virol. 2000;74:3321–3329. doi: 10.1128/JVI.74.7.3321-3329.2000. - DOI - PMC - PubMed
-
- Mangeney M., Renard M., Schlecht-Louf G., Bouallaga I., Heidmann O., Letzelter C., Richaud A., Ducos B., Heidmann T. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl. Acad. Sci. USA. 2007;104:20534–20539. doi: 10.1073/pnas.0707873105. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

