Current Practice in Bicistronic IRES Reporter Use: A Systematic Review
- PMID: 34068921
- PMCID: PMC8156625
- DOI: 10.3390/ijms22105193
Current Practice in Bicistronic IRES Reporter Use: A Systematic Review
Abstract
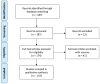
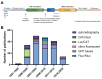
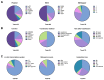
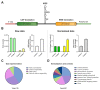
Bicistronic reporter assays have been instrumental for transgene expression, understanding of internal ribosomal entry site (IRES) translation, and identification of novel cap-independent translational elements (CITE). We observed a large methodological variability in the use of bicistronic reporter assays and data presentation or normalization procedures. Therefore, we systematically searched the literature for bicistronic IRES reporter studies and analyzed methodological details, data visualization, and normalization procedures. Two hundred fifty-seven publications were identified using our search strategy (published 1994-2020). Experimental studies on eukaryotic adherent cell systems and the cell-free translation assay were included for further analysis. We evaluated the following methodological details for 176 full text articles: the bicistronic reporter design, the cell line or type, transfection methods, and time point of analyses post-transfection. For the cell-free translation assay, we focused on methods of in vitro transcription, type of translation lysate, and incubation times and assay temperature. Data can be presented in multiple ways: raw data from individual cistrons, a ratio of the two, or fold changes thereof. In addition, many different control experiments have been suggested when studying IRES-mediated translation. In addition, many different normalization and control experiments have been suggested when studying IRES-mediated translation. Therefore, we also categorized and summarized their use. Our unbiased analyses provide a representative overview of bicistronic IRES reporter use. We identified parameters that were reported inconsistently or incompletely, which could hamper data reproduction and interpretation. On the basis of our analyses, we encourage adhering to a number of practices that should improve transparency of bicistronic reporter data presentation and improve methodological descriptions to facilitate data replication.
Keywords: IRES; bicistronic reporter; dicistronic reporter; mRNA translation; ribosome; systematic review.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs.Nucleic Acids Res. 2001 Aug 15;29(16):3327-34. doi: 10.1093/nar/29.16.3327. Nucleic Acids Res. 2001. PMID: 11504870 Free PMC article.
-
Differential bicistronic gene translation mediated by the internal ribosome entry site element of encephalomyocarditis virus.Biomed J. 2021 Dec;44(6 Suppl 1):S54-S62. doi: 10.1016/j.bj.2020.06.006. Epub 2020 Jun 24. Biomed J. 2021. PMID: 35747995 Free PMC article.
-
In vitro characterization of an internal ribosomal entry site (IRES) present within the 5' nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus.J Virol. 1994 Feb;68(2):1066-74. doi: 10.1128/JVI.68.2.1066-1074.1994. J Virol. 1994. PMID: 8289336 Free PMC article.
-
A researcher's guide to the galaxy of IRESs.Cell Mol Life Sci. 2017 Apr;74(8):1431-1455. doi: 10.1007/s00018-016-2409-5. Epub 2016 Nov 16. Cell Mol Life Sci. 2017. PMID: 27853833 Free PMC article. Review.
-
IRES-mediated cap-independent translation, a path leading to hidden proteome.J Mol Cell Biol. 2019 Oct 25;11(10):911-919. doi: 10.1093/jmcb/mjz091. J Mol Cell Biol. 2019. PMID: 31504667 Free PMC article. Review.
Cited by
-
Internal ribosomal entry site-mediated translational activity of nitric oxide synthase 2.Anim Cells Syst (Seoul). 2023 Nov 14;27(1):321-328. doi: 10.1080/19768354.2023.2275613. eCollection 2023. Anim Cells Syst (Seoul). 2023. PMID: 38414531 Free PMC article.
-
Validating the EMCV IRES Secondary Structure with Structure-Function Analysis.Biochemistry. 2024 Jan 2;63(1):107-115. doi: 10.1021/acs.biochem.3c00579. Epub 2023 Dec 11. Biochemistry. 2024. PMID: 38081770
-
The intrinsically disordered region of eIF5B stimulates IRES usage and nucleates biological granule formation.Cell Rep. 2023 Oct 31;42(10):113283. doi: 10.1016/j.celrep.2023.113283. Epub 2023 Oct 19. Cell Rep. 2023. PMID: 37862172 Free PMC article.
-
Integrative solution structure of PTBP1-IRES complex reveals strong compaction and ordering with residual conformational flexibility.Nat Commun. 2023 Oct 13;14(1):6429. doi: 10.1038/s41467-023-42012-z. Nat Commun. 2023. PMID: 37833274 Free PMC article.
-
Small Molecules Targeting Viral RNA.Int J Mol Sci. 2023 Aug 31;24(17):13500. doi: 10.3390/ijms241713500. Int J Mol Sci. 2023. PMID: 37686306 Free PMC article. Review.
References
-
- Jing Y., Simmer P., Feng J.Q. Cell Lineage Tracing: Colocalization of Cell Lineage Markers with a Fluorescent Reporter. Methods Mol. Biol. 2021;2230:325–335. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

