Hamster Polyomavirus Research: Past, Present, and Future
- PMID: 34068409
- PMCID: PMC8153644
- DOI: 10.3390/v13050907
Hamster Polyomavirus Research: Past, Present, and Future
Abstract
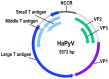
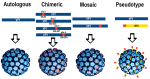
Hamster polyomavirus (Mesocricetus auratus polyomavirus 1, HaPyV) was discovered as one of the first rodent polyomaviruses at the end of the 1960s in a colony of Syrian hamsters (Mesocricetus auratus) affected by skin tumors. Natural HaPyV infections have been recorded in Syrian hamster colonies due to the occurrence of skin tumors and lymphomas. HaPyV infections of Syrian hamsters represent an important and pioneering tumor model. Experimental infections of Syrian hamsters of different colonies are still serving as model systems (e.g., mesothelioma). The observed phylogenetic relationship of HaPyV to murine polyomaviruses within the genus Alphapolyomavirus, and the exclusive detection of other cricetid polyomaviruses, i.e., common vole (Microtus arvalis polyomavirus 1) and bank vole (Myodes glareolus polyomavirus 1) polyomaviruses, in the genus Betapolyomavirus, must be considered with caution, as knowledge of rodent-associated polyomaviruses is still limited. The genome of HaPyV shows the typical organization of polyomaviruses with an early and a late transcriptional region. The early region encodes three tumor (T) antigens including a middle T antigen; the late region encodes three capsid proteins. The major capsid protein VP1 of HaPyV was established as a carrier for the generation of autologous, chimeric, and mosaic virus-like particles (VLPs) with a broad range of applications, e.g., for the production of epitope-specific antibodies. Autologous VLPs have been applied for entry and maturation studies of dendritic cells. The generation of chimeric and mosaic VLPs indicated the high flexibility of the VP1 carrier protein for the insertion of foreign sequences. The generation of pseudotype VLPs of original VP1 and VP2-foreign protein fusion can further enhance the applicability of this system. Future investigations should evaluate the evolutionary origin of HaPyV, monitor its occurrence in wildlife and Syrian hamster breeding, and prove its value for the generation of potential vaccine candidates and as a gene therapy vehicle.
Keywords: Syrian hamster; genome organization; major capsid protein; middle T antigen; rodent polyomaviruses; tumor model; virus discovery; virus-like particles.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Hamster polyomavirus-associated T-cell lymphomas in Syrian hamsters (Mesocricetus auratus).Vet Pathol. 2023 Mar;60(2):267-275. doi: 10.1177/03009858221140823. Epub 2022 Dec 20. Vet Pathol. 2023. PMID: 36537739
-
The hamster polyomavirus--a brief review of recent knowledge.Virus Genes. 2001 Jan;22(1):93-101. doi: 10.1023/a:1008190504521. Virus Genes. 2001. PMID: 11210944 Review.
-
Lymphoma outbreak in a GASH:Sal hamster colony.Arch Virol. 2013 Nov;158(11):2255-65. doi: 10.1007/s00705-013-1737-0. Epub 2013 May 30. Arch Virol. 2013. PMID: 23719671
-
Construction of polyomavirus-derived pseudotype virus-like particles displaying a functionally active neutralizing antibody against hepatitis B virus surface antigen.BMC Biotechnol. 2015 Sep 15;15:85. doi: 10.1186/s12896-015-0203-3. BMC Biotechnol. 2015. PMID: 26370129 Free PMC article.
-
Production and biomedical applications of virus-like particles derived from polyomaviruses.J Control Release. 2013 Nov 28;172(1):305-321. doi: 10.1016/j.jconrel.2013.08.026. Epub 2013 Aug 31. J Control Release. 2013. PMID: 23999392 Review.
Cited by
-
Functional Domains of the Early Proteins and Experimental and Epidemiological Studies Suggest a Role for the Novel Human Polyomaviruses in Cancer.Front Microbiol. 2022 Feb 18;13:834368. doi: 10.3389/fmicb.2022.834368. eCollection 2022. Front Microbiol. 2022. PMID: 35250950 Free PMC article. Review.
-
Vaccination with Prion Peptide-Displaying Polyomavirus-Like Particles Prolongs Incubation Time in Scrapie-Infected Mice.Viruses. 2021 Apr 30;13(5):811. doi: 10.3390/v13050811. Viruses. 2021. PMID: 33946367 Free PMC article.
-
Characterization of the gastrointestinal microbiome of the Syrian hamster (Mesocricetus auratus) and comparison to data from mice.FEBS Open Bio. 2024 Oct;14(10):1701-1717. doi: 10.1002/2211-5463.13869. Epub 2024 Aug 4. FEBS Open Bio. 2024. PMID: 39097990 Free PMC article.
-
Golden Syrian Hamster Models for Cancer Research.Cells. 2022 Aug 3;11(15):2395. doi: 10.3390/cells11152395. Cells. 2022. PMID: 35954238 Free PMC article. Review.
References
-
- DeCaprio J.A., Imperiale M.J., Major E.O. Polyomaviruses. In: Knipe D.M., Howley P.W., editors. Fields Virology. 6th ed. Wolters Kluwer; Alphen aan den Rijn, The Netherlands: 2013. pp. 1633–1661.
-
- Ehlers B., Anoh A.E., Ben Salem N., Broll S., Couacy-Hymann E., Fischer D., Gedvilaite A., Ingenhutt N., Liebmann S., Martin M., et al. Novel Polyomaviruses in Mammals from Multiple Orders and Reassessment of Polyomavirus Evolution and Taxonomy. Viruses. 2019;11:930. doi: 10.3390/v11100930. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

