The Bunyavirales: The Plant-Infecting Counterparts
- PMID: 34066457
- PMCID: PMC8148189
- DOI: 10.3390/v13050842
The Bunyavirales: The Plant-Infecting Counterparts
Abstract
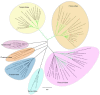
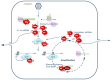
Negative-strand (-) RNA viruses (NSVs) comprise a large and diverse group of viruses that are generally divided in those with non-segmented and those with segmented genomes. Whereas most NSVs infect animals and humans, the smaller group of the plant-infecting counterparts is expanding, with many causing devastating diseases worldwide, affecting a large number of major bulk and high-value food crops. In 2018, the taxonomy of segmented NSVs faced a major reorganization with the establishment of the order Bunyavirales. This article overviews the major plant viruses that are part of the order, i.e., orthospoviruses (Tospoviridae), tenuiviruses (Phenuiviridae), and emaraviruses (Fimoviridae), and provides updates on the more recent ongoing research. Features shared with the animal-infecting counterparts are mentioned, however, special attention is given to their adaptation to plant hosts and vector transmission, including intra/intercellular trafficking and viral counter defense to antiviral RNAi.
Keywords: EMARaV; Emaravirus; European mountain ash ringspot-associated virus; Fimoviridae; Orthotospovirus; Phenuiviridae; RSV; Rice stripe virus; TSWV; Tenuivirus; Tomato spotted wilt virus; Tospoviridae.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Negative-strand RNA viruses: the plant-infecting counterparts.Virus Res. 2011 Dec;162(1-2):184-202. doi: 10.1016/j.virusres.2011.09.028. Epub 2011 Sep 22. Virus Res. 2011. PMID: 21963660 Review.
-
Family Level Phylogenies Reveal Relationships of Plant Viruses within the Order Bunyavirales.Viruses. 2020 Sep 10;12(9):1010. doi: 10.3390/v12091010. Viruses. 2020. PMID: 32927652 Free PMC article.
-
ICTV Virus Taxonomy Profile: Fimoviridae.J Gen Virol. 2018 Nov;99(11):1478-1479. doi: 10.1099/jgv.0.001143. Epub 2018 Sep 11. J Gen Virol. 2018. PMID: 30204080
-
Emaravirus-specific degenerate PCR primers allowed the identification of partial RNA-dependent RNA polymerase sequences of Maize red stripe virus and Pigeonpea sterility mosaic virus.J Virol Methods. 2013 Mar;188(1-2):37-40. doi: 10.1016/j.jviromet.2012.11.037. Epub 2012 Dec 7. J Virol Methods. 2013. PMID: 23219928
-
Emaravirus: a novel genus of multipartite, negative strand RNA plant viruses.Viruses. 2012 Sep;4(9):1515-36. doi: 10.3390/v4091515. Epub 2012 Sep 12. Viruses. 2012. PMID: 23170170 Free PMC article. Review.
Cited by
-
Mixed infection of an emaravirus, a crinivirus, and a begomovirus in Pueraria lobata (Willd) Ohwi.Front Microbiol. 2022 Sep 29;13:926724. doi: 10.3389/fmicb.2022.926724. eCollection 2022. Front Microbiol. 2022. PMID: 36246248 Free PMC article.
-
Assembly of plant virus agroinfectious clones using biological material or DNA synthesis.STAR Protoc. 2022 Dec 16;3(4):101716. doi: 10.1016/j.xpro.2022.101716. Epub 2022 Sep 22. STAR Protoc. 2022. PMID: 36149792 Free PMC article.
-
The Plant Virus Tomato Spotted Wilt Orthotospovirus Benefits Its Vector Frankliniella occidentalis by Decreasing Plant Toxic Alkaloids in Host Plant Datura stramonium.Int J Mol Sci. 2023 Sep 24;24(19):14493. doi: 10.3390/ijms241914493. Int J Mol Sci. 2023. PMID: 37833941 Free PMC article.
-
Determination of key residues in tospoviral NSm required for Sw-5b recognition, their potential ability to overcome resistance, and the effective resistance provided by improved Sw-5b mutants.Mol Plant Pathol. 2022 May;23(5):622-633. doi: 10.1111/mpp.13182. Epub 2021 Dec 27. Mol Plant Pathol. 2022. PMID: 34962031 Free PMC article.
-
Autophagy plays an antiviral defence role against tomato spotted wilt orthotospovirus and is counteracted by viral effector NSs.Mol Plant Pathol. 2024 Oct;25(10):e70012. doi: 10.1111/mpp.70012. Mol Plant Pathol. 2024. PMID: 39350560 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

