An Integrated Approach toward NanoBRET Tracers for Analysis of GPCR Ligand Engagement
- PMID: 34065854
- PMCID: PMC8151276
- DOI: 10.3390/molecules26102857
An Integrated Approach toward NanoBRET Tracers for Analysis of GPCR Ligand Engagement
Abstract
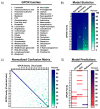
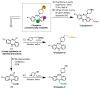
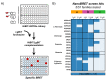
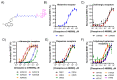
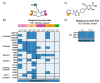
Gaining insight into the pharmacology of ligand engagement with G-protein coupled receptors (GPCRs) under biologically relevant conditions is vital to both drug discovery and basic research. NanoLuc-based bioluminescence resonance energy transfer (NanoBRET) monitoring competitive binding between fluorescent tracers and unmodified test compounds has emerged as a robust and sensitive method to quantify ligand engagement with specific GPCRs genetically fused to NanoLuc luciferase or the luminogenic HiBiT peptide. However, development of fluorescent tracers is often challenging and remains the principal bottleneck for this approach. One way to alleviate the burden of developing a specific tracer for each receptor is using promiscuous tracers, which is made possible by the intrinsic specificity of BRET. Here, we devised an integrated tracer discovery workflow that couples machine learning-guided in silico screening for scaffolds displaying promiscuous binding to GPCRs with a blend of synthetic strategies to rapidly generate multiple tracer candidates. Subsequently, these candidates were evaluated for binding in a NanoBRET ligand-engagement screen across a library of HiBiT-tagged GPCRs. Employing this workflow, we generated several promiscuous fluorescent tracers that can effectively engage multiple GPCRs, demonstrating the efficiency of this approach. We believe that this workflow has the potential to accelerate discovery of NanoBRET fluorescent tracers for GPCRs and other target classes.
Keywords: GPCRS; HiBiT; NanoBRET; in silico screen; ligand-engagement.
Conflict of interest statement
All authors were Promega employees at time of manuscript preparation. The authors declare no conflict of interest.
Figures


Similar articles
-
Luminogenic HiBiT Peptide-Based NanoBRET Ligand Binding Assays for Melatonin Receptors.ACS Pharmacol Transl Sci. 2022 Jul 18;5(8):668-678. doi: 10.1021/acsptsci.2c00096. eCollection 2022 Aug 12. ACS Pharmacol Transl Sci. 2022. PMID: 35983281 Free PMC article.
-
The luminescent HiBiT peptide enables selective quantitation of G protein-coupled receptor ligand engagement and internalization in living cells.J Biol Chem. 2020 Apr 10;295(15):5124-5135. doi: 10.1074/jbc.RA119.011952. Epub 2020 Feb 27. J Biol Chem. 2020. PMID: 32107310 Free PMC article.
-
NanoBRET Approaches to Study Ligand Binding to GPCRs and RTKs.Trends Pharmacol Sci. 2018 Feb;39(2):136-147. doi: 10.1016/j.tips.2017.10.006. Epub 2017 Nov 10. Trends Pharmacol Sci. 2018. PMID: 29132917 Review.
-
New Horizons on Molecular Pharmacology Applied to Drug Discovery: When Resonance Overcomes Radioligand Binding.Curr Radiopharm. 2017;10(1):16-20. doi: 10.2174/1874471010666170208152420. Curr Radiopharm. 2017. PMID: 28183248 Review.
-
Equilibrium and Kinetic Measurements of Ligand Binding to HiBiT-tagged GPCRs on the Surface of Living Cells.Bio Protoc. 2020 Dec 20;10(24):e3861. doi: 10.21769/BioProtoc.3861. eCollection 2020 Dec 20. Bio Protoc. 2020. PMID: 33659503 Free PMC article.
Cited by
-
Luminogenic HiBiT Peptide-Based NanoBRET Ligand Binding Assays for Melatonin Receptors.ACS Pharmacol Transl Sci. 2022 Jul 18;5(8):668-678. doi: 10.1021/acsptsci.2c00096. eCollection 2022 Aug 12. ACS Pharmacol Transl Sci. 2022. PMID: 35983281 Free PMC article.
-
Advances in luminescence-based technologies for drug discovery.Expert Opin Drug Discov. 2023 Jan;18(1):25-35. doi: 10.1080/17460441.2023.2160441. Epub 2022 Dec 23. Expert Opin Drug Discov. 2023. PMID: 36562206 Free PMC article. Review.
-
N-(4-Bromo-2,5-Dimethoxyphenethyl)-6-(4-Phenylbutoxy)Hexan-1-Amine (XOB): A Novel Phenylalkylamine Antagonist of Serotonin 2A Receptors and Voltage-Gated Sodium Channels.Mol Pharmacol. 2024 Jul 17;106(2):92-106. doi: 10.1124/molpharm.123.000837. Mol Pharmacol. 2024. PMID: 38821630
-
Genome-wide pan-GPCR cell libraries accelerate drug discovery.Acta Pharm Sin B. 2024 Oct;14(10):4296-4311. doi: 10.1016/j.apsb.2024.06.023. Epub 2024 Jun 26. Acta Pharm Sin B. 2024. PMID: 39525595 Free PMC article. Review.
References
-
- Boursier M.E., Levin S., Zimmerman K., Machleidt T., Hurst R., Butler B.L., Eggers C.T., Kirkland T.A., Wood K.V., Ohana R.F. The luminescent HiBiT peptide enables selective quantitation of G protein–coupled receptor ligand engagement and internalization in living cells. J. Biol. Chem. 2020;295:5124–5135. doi: 10.1074/jbc.RA119.011952. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

