How DNA and RNA Viruses Exploit Host Chaperones to Promote Infection
- PMID: 34064125
- PMCID: PMC8224278
- DOI: 10.3390/v13060958
How DNA and RNA Viruses Exploit Host Chaperones to Promote Infection
Abstract
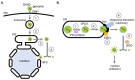
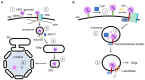
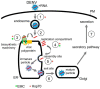
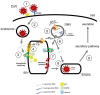
To initiate infection, a virus enters a host cell typically via receptor-dependent endocytosis. It then penetrates a subcellular membrane, reaching a destination that supports transcription, translation, and replication of the viral genome. These steps lead to assembly and morphogenesis of the new viral progeny. The mature virus finally exits the host cell to begin the next infection cycle. Strikingly, viruses hijack host molecular chaperones to accomplish these distinct entry steps. Here we highlight how DNA viruses, including polyomavirus and the human papillomavirus, exploit soluble and membrane-associated chaperones to enter a cell, penetrating and escaping an intracellular membrane en route for infection. We also describe the mechanism by which RNA viruses-including flavivirus and coronavirus-co-opt cytosolic and organelle-selective chaperones to promote viral endocytosis, protein biosynthesis, replication, and assembly. These examples underscore the importance of host chaperones during virus infection, potentially revealing novel antiviral strategies to combat virus-induced diseases.
Keywords: Golgi; chaperones; coronavirus; endoplasmic reticulum; flavivirus; human papillomavirus; infection; polyomavirus SV40; viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
ER functions are exploited by viruses to support distinct stages of their life cycle.Biochem Soc Trans. 2020 Oct 30;48(5):2173-2184. doi: 10.1042/BST20200395. Biochem Soc Trans. 2020. PMID: 33119046 Free PMC article. Review.
-
How non-enveloped viruses hijack host machineries to cause infection.Adv Virus Res. 2019;104:97-122. doi: 10.1016/bs.aivir.2019.05.002. Epub 2019 Jul 2. Adv Virus Res. 2019. PMID: 31439154 Free PMC article.
-
SGTA-Dependent Regulation of Hsc70 Promotes Cytosol Entry of Simian Virus 40 from the Endoplasmic Reticulum.J Virol. 2017 May 26;91(12):e00232-17. doi: 10.1128/JVI.00232-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28356524 Free PMC article.
-
How viruses use the endoplasmic reticulum for entry, replication, and assembly.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a013250. doi: 10.1101/cshperspect.a013250. Cold Spring Harb Perspect Biol. 2013. PMID: 23284050 Free PMC article. Review.
-
How host ER membrane chaperones and morphogenic proteins support virus infection.J Cell Sci. 2023 Jul 1;136(13):jcs261121. doi: 10.1242/jcs.261121. Epub 2023 Jul 4. J Cell Sci. 2023. PMID: 37401530 Free PMC article.
Cited by
-
Non-enveloped virus membrane penetration: New advances leading to new insights.PLoS Pathog. 2022 Dec 8;18(12):e1010948. doi: 10.1371/journal.ppat.1010948. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36480535 Free PMC article.
-
Reticulons promote formation of ER-derived double-membrane vesicles that facilitate SARS-CoV-2 replication.J Cell Biol. 2023 Jul 3;222(7):e202203060. doi: 10.1083/jcb.202203060. Epub 2023 Apr 24. J Cell Biol. 2023. PMID: 37093123 Free PMC article.
-
Carbohydrates Metabolic Signatures in Immune Cells: Response to Infection.Front Immunol. 2022 Jul 4;13:912899. doi: 10.3389/fimmu.2022.912899. eCollection 2022. Front Immunol. 2022. PMID: 35983037 Free PMC article.
-
Autophagy of the ER: the secretome finds the lysosome.FEBS J. 2023 Dec;290(24):5656-5673. doi: 10.1111/febs.16986. Epub 2023 Nov 3. FEBS J. 2023. PMID: 37920925 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

