Molecular Insights into the Flavivirus Replication Complex
- PMID: 34064113
- PMCID: PMC8224304
- DOI: 10.3390/v13060956
Molecular Insights into the Flavivirus Replication Complex
Abstract
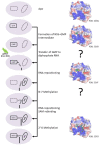
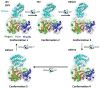
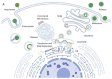
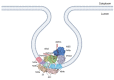
Flaviviruses are vector-borne RNA viruses, many of which are clinically relevant human viral pathogens, such as dengue, Zika, Japanese encephalitis, West Nile and yellow fever viruses. Millions of people are infected with these viruses around the world each year. Vaccines are only available for some members of this large virus family, and there are no effective antiviral drugs to treat flavivirus infections. The unmet need for vaccines and therapies against these flaviviral infections drives research towards a better understanding of the epidemiology, biology and immunology of flaviviruses. In this review, we discuss the basic biology of the flavivirus replication process and focus on the molecular aspects of viral genome replication. Within the virus-induced intracellular membranous compartments, flaviviral RNA genome replication takes place, starting from viral poly protein expression and processing to the assembly of the virus RNA replication complex, followed by the delivery of the progeny viral RNA to the viral particle assembly sites. We attempt to update the latest understanding of the key molecular events during this process and highlight knowledge gaps for future studies.
Keywords: RNA replication; flavivirus; non-structural protein; replication complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Role of RNA-binding proteins during the late stages of Flavivirus replication cycle.Virol J. 2020 Apr 25;17(1):60. doi: 10.1186/s12985-020-01329-7. Virol J. 2020. PMID: 32334603 Free PMC article. Review.
-
RPLP1 and RPLP2 Are Essential Flavivirus Host Factors That Promote Early Viral Protein Accumulation.J Virol. 2017 Jan 31;91(4):e01706-16. doi: 10.1128/JVI.01706-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27974556 Free PMC article.
-
Zinc Finger Protein ZFP36L1 Inhibits Flavivirus Infection by both 5'-3' XRN1 and 3'-5' RNA-Exosome RNA Decay Pathways.J Virol. 2022 Jan 12;96(1):e0166521. doi: 10.1128/JVI.01665-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643435 Free PMC article.
-
The Host Factor AUF1 p45 Supports Flavivirus Propagation by Triggering the RNA Switch Required for Viral Genome Cyclization.J Virol. 2018 Feb 26;92(6):e01647-17. doi: 10.1128/JVI.01647-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29263261 Free PMC article.
-
The Many Faces of the Flavivirus NS5 Protein in Antagonism of Type I Interferon Signaling.J Virol. 2017 Jan 18;91(3):e01970-16. doi: 10.1128/JVI.01970-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881649 Free PMC article. Review.
Cited by
-
RNA virus infections and their effect on host alternative splicing.Antiviral Res. 2023 Feb;210:105503. doi: 10.1016/j.antiviral.2022.105503. Epub 2022 Dec 23. Antiviral Res. 2023. PMID: 36572191 Free PMC article. Review.
-
Flavivirus nonstructural proteins and replication complexes as antiviral drug targets.Curr Opin Virol. 2023 Apr;59:101305. doi: 10.1016/j.coviro.2023.101305. Epub 2023 Mar 2. Curr Opin Virol. 2023. PMID: 36870091 Free PMC article. Review.
-
2'-O-methyltransferase-deficient yellow fever virus: Restricted replication in the midgut and secondary tissues of Aedes aegypti mosquitoes severely limits dissemination.PLoS Pathog. 2024 Oct 2;20(10):e1012607. doi: 10.1371/journal.ppat.1012607. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39356716 Free PMC article.
-
Flavivirus NS4B protein: Structure, function, and antiviral discovery.Antiviral Res. 2022 Nov;207:105423. doi: 10.1016/j.antiviral.2022.105423. Epub 2022 Sep 27. Antiviral Res. 2022. PMID: 36179934 Free PMC article. Review.
-
Combined NMR and molecular dynamics conformational filter identifies unambiguously dynamic ensembles of Dengue protease NS2B/NS3pro.Commun Biol. 2023 Nov 24;6(1):1193. doi: 10.1038/s42003-023-05584-6. Commun Biol. 2023. PMID: 38001280 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

