A Combined Analysis of Gut and Skin Microbiota in Infants with Food Allergy and Atopic Dermatitis: A Pilot Study
- PMID: 34063398
- PMCID: PMC8156695
- DOI: 10.3390/nu13051682
A Combined Analysis of Gut and Skin Microbiota in Infants with Food Allergy and Atopic Dermatitis: A Pilot Study
Abstract
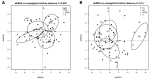
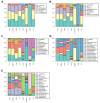
The gut microbiota in patients with food allergy, and the skin microbiota in atopic dermatitis patients differ from those of healthy people. We hypothesize that relationships may exist between gut and skin microbiota in patients with allergies. The aim of this study was to determine the possible relationship between gut and skin microbiota in patients with allergies, hence simultaneous analysis of the two compartments of microbiota was performed in infants with and without allergic symptoms. Fifty-nine infants with food allergy and/or atopic dermatitis and 28 healthy children were enrolled in the study. The skin and gut microbiota were evaluated using 16S rRNA gene amplicon sequencing. No significant differences in the α-diversity of dermal or fecal microbiota were observed between allergic and non-allergic infants; however, a significant relationship was found between bacterial community structure and allergy phenotypes, especially in the fecal samples. Certain clinical conditions were associated with characteristic bacterial taxa in the skin and gut microbiota. Positive correlations were found between skin and fecal samples in the abundance of Gemella among allergic infants, and Lactobacillus and Bacteroides among healthy infants. Although infants with allergies and healthy infants demonstrate microbiota with similar α-diversity, some differences in β-diversity and bacterial species abundance can be seen, which may depend on the phenotype of the allergy. For some organisms, their abundance in skin and feces samples may be correlated, and these correlations might serve as indicators of the host's allergic state.
Keywords: 16S rRNA sequencing; atopic dermatitis; dysbiosis; food allergy; gut; infants; microbiota; skin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Mother's Milk Microbiome Shaping Fecal and Skin Microbiota in Infants with Food Allergy and Atopic Dermatitis: A Pilot Analysis.Nutrients. 2021 Oct 14;13(10):3600. doi: 10.3390/nu13103600. Nutrients. 2021. PMID: 34684601 Free PMC article.
-
Fecal Microbiome and Food Allergy in Pediatric Atopic Dermatitis: A Cross-Sectional Pilot Study.Int Arch Allergy Immunol. 2018;175(1-2):77-84. doi: 10.1159/000484897. Epub 2018 Jan 25. Int Arch Allergy Immunol. 2018. PMID: 29393195
-
Differences in gut microbiota between allergic rhinitis, atopic dermatitis, and skin urticaria: A pilot study.Medicine (Baltimore). 2021 Mar 5;100(9):e25091. doi: 10.1097/MD.0000000000025091. Medicine (Baltimore). 2021. PMID: 33655988 Free PMC article.
-
Demystifying Dysbiosis: Can the Gut Microbiome Promote Oral Tolerance Over IgE-mediated Food Allergy?Curr Pediatr Rev. 2018;14(3):156-163. doi: 10.2174/1573396314666180507120424. Curr Pediatr Rev. 2018. PMID: 29732975 Review.
-
Recent developments and highlights in mechanisms of allergic diseases: Microbiome.Allergy. 2018 Dec;73(12):2314-2327. doi: 10.1111/all.13634. Epub 2018 Nov 13. Allergy. 2018. PMID: 30325537 Review.
Cited by
-
Atopic dermatitis is associated with abnormal stool form: a population-based cross-sectional study in college students.Arch Dermatol Res. 2023 Sep;315(7):2057-2064. doi: 10.1007/s00403-023-02567-9. Epub 2023 Mar 14. Arch Dermatol Res. 2023. PMID: 36917250
-
Lipidomics and Metabolomics in Infant Atopic Dermatitis: What's the Correlation with Early Nutrition?Curr Pediatr Rev. 2024;20(4):510-524. doi: 10.2174/1573396320666230411093122. Curr Pediatr Rev. 2024. PMID: 37055903 Review.
-
Gender Differences in the Interplay between Vitamin D and Microbiota in Allergic and Autoimmune Diseases.Biomedicines. 2024 May 7;12(5):1023. doi: 10.3390/biomedicines12051023. Biomedicines. 2024. PMID: 38790985 Free PMC article. Review.
-
Attenuation of allergen-specific immunotherapy for atopic dermatitis by ectopic colonization of Brevundimonas vesicularis in the intestine.Cell Rep Med. 2023 Dec 19;4(12):101340. doi: 10.1016/j.xcrm.2023.101340. Cell Rep Med. 2023. PMID: 38118418 Free PMC article.
-
Mother's Milk Microbiome Shaping Fecal and Skin Microbiota in Infants with Food Allergy and Atopic Dermatitis: A Pilot Analysis.Nutrients. 2021 Oct 14;13(10):3600. doi: 10.3390/nu13103600. Nutrients. 2021. PMID: 34684601 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

