Dynamics of Proteins and Macromolecular Machines in Escherichia coli
- PMID: 34060908
- PMCID: PMC11163846
- DOI: 10.1128/ecosalplus.ESP-0011-2020
Dynamics of Proteins and Macromolecular Machines in Escherichia coli
Abstract
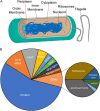
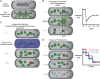
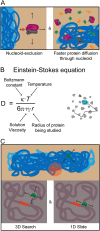
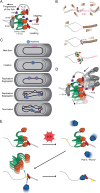
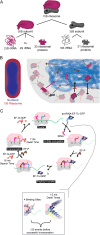
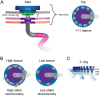
Proteins are major contributors to the composition and the functions in the cell. They often assemble into larger structures, macromolecular machines, to carry out intricate essential functions. Although huge progress in understanding how macromolecular machines function has been made by reconstituting them in vitro, the role of the intracellular environment is still emerging. The development of fluorescence microscopy techniques in the last 2 decades has allowed us to obtain an increased understanding of proteins and macromolecular machines in cells. Here, we describe how proteins move by diffusion, how they search for their targets, and how they are affected by the intracellular environment. We also describe how proteins assemble into macromolecular machines and provide examples of how frequent subunit turnover is used for them to function and to respond to changes in the intracellular conditions. This review emphasizes the constant movement of molecules in cells, the stochastic nature of reactions, and the dynamic nature of macromolecular machines.
Keywords: DNA replication; flagellar motor; fluorescence microscopy; molecular machine; protein diffusion; protein dynamics; protein target search; replisome; ribosome.
Figures
Similar articles
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Stoichiometry and turnover of the bacterial flagellar switch protein FliN.mBio. 2014 Jul 1;5(4):e01216-14. doi: 10.1128/mBio.01216-14. mBio. 2014. PMID: 24987089 Free PMC article.
-
Stoichiometry and architecture of active DNA replication machinery in Escherichia coli.Science. 2010 Apr 23;328(5977):498-501. doi: 10.1126/science.1185757. Science. 2010. PMID: 20413500 Free PMC article.
-
The Macromolecular Machines that Duplicate the Escherichia coli Chromosome as Targets for Drug Discovery.Antibiotics (Basel). 2018 Mar 14;7(1):23. doi: 10.3390/antibiotics7010023. Antibiotics (Basel). 2018. PMID: 29538288 Free PMC article. Review.
-
A quest for coordination among activities at the replisome.Biochem Soc Trans. 2019 Aug 30;47(4):1067-1075. doi: 10.1042/BST20180402. Epub 2019 Aug 8. Biochem Soc Trans. 2019. PMID: 31395754 Review.
References
-
- Hu P, Janga SC, Babu M, Díaz-Mejía JJ, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, Chandran S, Christopoulos C, Nazarians-Armavil A, Nasseri NK, Musso G, Ali M, Nazemof N, Eroukova V, Golshani A, Paccanaro A, Greenblatt JF, Moreno-Hagelsieb G, Emili A. 2009. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol 7:e1000096. doi:10.1371/journal.pbio.1000096. - DOI - PMC - PubMed
-
- Neidhardt FCN, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell: a molecular approach.article-title. Sinauer Associates, Sunderland, MA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

