Resolution of Low-Energy States in Spin-Exchange Transition-Metal Clusters: Case Study of Singlet States in [Fe(III)4S4] Cubanes
- PMID: 34048648
- PMCID: PMC8201447
- DOI: 10.1021/acs.jpca.1c00397
Resolution of Low-Energy States in Spin-Exchange Transition-Metal Clusters: Case Study of Singlet States in [Fe(III)4S4] Cubanes
Abstract
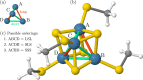
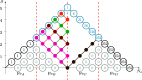
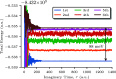
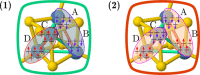
Polynuclear transition-metal (PNTM) clusters owe their catalytic activity to numerous energetically low-lying spin states and stable oxidation states. The characterization of their electronic structure represents one of the greatest challenges of modern chemistry. We propose a theoretical framework that enables the resolution of targeted electronic states with ease and apply it to two [Fe(III)4S4] cubanes. Through direct access to their many-body wave functions, we identify important correlation mechanisms and their interplay with the geometrical distortions observed in these clusters, which are core properties in understanding their catalytic activity. The simulated magnetic coupling constants predicted by our strategy allow us to make qualitative connections between spin interactions and geometrical distortions, demonstrating its predictive power. Moreover, despite its simplicity, the strategy provides magnetic coupling constants in good agreement with the available experimental ones. The complexes are intrinsically frustrated anti-ferromagnets, and the obtained spin structures together with the geometrical distortions represent two possible ways to release spin frustration (spin-driven Jahn-Teller distortion). Our paradigm provides a simple, yet rigorous, route to uncover the electronic structure of PNTM clusters and may be applied to a wide variety of such clusters.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Modeling magnetic interactions in high-valent trinuclear [Mn3(IV)O4]4+ complexes through highly compressed multi-configurational wave functions.Phys Chem Chem Phys. 2021 Sep 15;23(35):19766-19780. doi: 10.1039/d1cp03259c. Phys Chem Chem Phys. 2021. PMID: 34525156
-
Oxidation state dependence of the geometry, electronic structure, and magnetic coupling in mixed oxo- and carboxylato-bridged manganese dimers.Inorg Chem. 2001 Jun 18;40(13):3061-76. doi: 10.1021/ic0008767. Inorg Chem. 2001. PMID: 11399174
-
Resolution of Electronic States in Heisenberg Cluster Models within the Unitary Group Approach.J Chem Theory Comput. 2023 Feb 28;19(4):1218-1230. doi: 10.1021/acs.jctc.2c01132. Epub 2023 Feb 3. J Chem Theory Comput. 2023. PMID: 36735906 Free PMC article.
-
DFT-based studies on the Jahn-Teller effect in 3d hexacyanometalates with orbitally degenerate ground states.J Phys Chem A. 2007 Sep 20;111(37):9145-63. doi: 10.1021/jp0731912. Epub 2007 Aug 24. J Phys Chem A. 2007. PMID: 17718456
-
Magnetic Metal Clusters and Superatoms.J Phys Chem Lett. 2024 Feb 22;15(7):1856-1865. doi: 10.1021/acs.jpclett.3c03637. Epub 2024 Feb 9. J Phys Chem Lett. 2024. PMID: 38335129 Review.
Cited by
-
Exploiting Locality in Full Configuration Interaction Quantum Monte Carlo for Fast Excitation Generation.J Chem Theory Comput. 2023 Dec 26;19(24):9118-9135. doi: 10.1021/acs.jctc.3c00546. Epub 2023 Dec 5. J Chem Theory Comput. 2023. PMID: 38051202 Free PMC article.
-
Spin-Pure Stochastic-CASSCF via GUGA-FCIQMC Applied to Iron-Sulfur Clusters.J Chem Theory Comput. 2021 Sep 14;17(9):5684-5703. doi: 10.1021/acs.jctc.1c00589. Epub 2021 Sep 1. J Chem Theory Comput. 2021. PMID: 34469685 Free PMC article.
-
Magnetic Interactions in a [Co(II)3Er(III)(OR)4] Model Cubane through Forefront Multiconfigurational Methods.J Chem Theory Comput. 2023 May 23;19(10):2811-2826. doi: 10.1021/acs.jctc.2c01318. Epub 2023 May 1. J Chem Theory Comput. 2023. PMID: 37126736 Free PMC article.
-
The energy landscape of magnetic materials.NPJ Comput Mater. 2024;10(1):151. doi: 10.1038/s41524-024-01310-w. Epub 2024 Jul 16. NPJ Comput Mater. 2024. PMID: 39026599 Free PMC article.
-
Analysis of the Geometric and Electronic Structure of Spin-Coupled Iron-Sulfur Dimers with Broken-Symmetry DFT: Implications for FeMoco.J Chem Theory Comput. 2022 Mar 8;18(3):1437-1457. doi: 10.1021/acs.jctc.1c00753. Epub 2022 Feb 15. J Chem Theory Comput. 2022. PMID: 35167749 Free PMC article.
References
-
- Renger G. Biological Exploitation of Solar Energy by Photosynthetic Water Splitting. Angew. Chem., Int. Ed. 1987, 26, 643–660. 10.1002/anie.198706431. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

