Regulation of host and viral promoters during human cytomegalovirus latency via US28 and CTCF
- PMID: 34042564
- PMCID: PMC8295918
- DOI: 10.1099/jgv.0.001609
Regulation of host and viral promoters during human cytomegalovirus latency via US28 and CTCF
Abstract
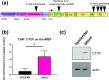
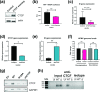
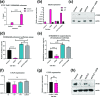
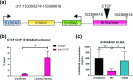
Viral latency is an active process during which the host cell environment is optimized for latent carriage and reactivation. This requires control of both viral and host gene promoters and enhancers often at the level of chromatin, and several viruses co-opt the chromatin organiser CTCF to control gene expression during latency. While CTCF has a role in the latencies of alpha- and gamma-herpesviruses, it was not known whether CTCF played a role in the latency of the beta-herpesvirus human cytomegalovirus (HCMV). Here, we show that HCMV latency is associated with increased CTCF expression and CTCF binding to the viral major lytic promoter, the major immediate early promoter (MIEP). This increase in CTCF binding is dependent on the virally encoded G protein coupled receptor, US28, and contributes to suppression of MIEP-driven transcription, a hallmark of latency. Furthermore, we show that latency-associated upregulation of CTCF represses expression of the neutrophil chemoattractants S100A8 and S100A9 which we have previously shown are downregulated during HCMV latency. As with downregulation of the MIEP, CTCF binding to the enhancer region of S100A8/A9 drives their suppression, again in a US28-dependent manner. Taken together, we identify CTCF upregulation as an important mechanism for optimizing latent carriage of HCMV at both the levels of viral and cellular gene expression.
Keywords: CTCF; chromatin; cytomegalovirus; latency; viral gene expression; virus-host interactions.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
Similar articles
-
Loopy virus or controlled contortionist? 3D regulation of HCMV gene expression by CTCF-driven chromatin interactions.J Virol. 2024 Oct 22;98(10):e0114824. doi: 10.1128/jvi.01148-24. Epub 2024 Aug 30. J Virol. 2024. PMID: 39212383 Review.
-
Host-encoded CTCF regulates human cytomegalovirus latency via chromatin looping.Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2315860121. doi: 10.1073/pnas.2315860121. Epub 2024 Feb 26. Proc Natl Acad Sci U S A. 2024. PMID: 38408244 Free PMC article.
-
Human cytomegalovirus G protein-coupled receptor US28 promotes latency by attenuating c-fos.Proc Natl Acad Sci U S A. 2019 Jan 29;116(5):1755-1764. doi: 10.1073/pnas.1816933116. Epub 2019 Jan 15. Proc Natl Acad Sci U S A. 2019. PMID: 30647114 Free PMC article.
-
Human Cytomegalovirus US28 Ligand Binding Activity Is Required for Latency in CD34+ Hematopoietic Progenitor Cells and Humanized NSG Mice.mBio. 2019 Aug 20;10(4):e01889-19. doi: 10.1128/mBio.01889-19. mBio. 2019. PMID: 31431555 Free PMC article.
-
HCMV latency: what regulates the regulators?Med Microbiol Immunol. 2019 Aug;208(3-4):431-438. doi: 10.1007/s00430-019-00581-1. Epub 2019 Feb 14. Med Microbiol Immunol. 2019. PMID: 30761409 Free PMC article. Review.
Cited by
-
Virus-Specific Nanobody-Chimeras Degrade the Human Cytomegalovirus US28 Protein in CD34+ Cells.Pathogens. 2024 Sep 24;13(10):821. doi: 10.3390/pathogens13100821. Pathogens. 2024. PMID: 39452693 Free PMC article.
-
Loopy virus or controlled contortionist? 3D regulation of HCMV gene expression by CTCF-driven chromatin interactions.J Virol. 2024 Oct 22;98(10):e0114824. doi: 10.1128/jvi.01148-24. Epub 2024 Aug 30. J Virol. 2024. PMID: 39212383 Review.
-
Delivery of US28 by incoming HCMV particles rapidly attenuates Akt activity to suppress HCMV lytic replication in monocytes.Sci Signal. 2024 Aug 27;17(851):eadn8727. doi: 10.1126/scisignal.adn8727. Epub 2024 Aug 27. Sci Signal. 2024. PMID: 39190708 Free PMC article.
-
cGAS-STING-TBK1 Signaling Promotes Valproic Acid-Responsive Human Cytomegalovirus Immediate-Early Transcription during Infection of Incompletely Differentiated Myeloid Cells.Viruses. 2024 May 30;16(6):877. doi: 10.3390/v16060877. Viruses. 2024. PMID: 38932169 Free PMC article.
-
Host-encoded CTCF regulates human cytomegalovirus latency via chromatin looping.Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2315860121. doi: 10.1073/pnas.2315860121. Epub 2024 Feb 26. Proc Natl Acad Sci U S A. 2024. PMID: 38408244 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

