Multimodally profiling memory T cells from a tuberculosis cohort identifies cell state associations with demographics, environment and disease
- PMID: 34031617
- PMCID: PMC8162307
- DOI: 10.1038/s41590-021-00933-1
Multimodally profiling memory T cells from a tuberculosis cohort identifies cell state associations with demographics, environment and disease
Abstract
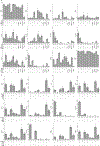
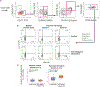
Multimodal T cell profiling can enable more precise characterization of elusive cell states underlying disease. Here, we integrated single-cell RNA and surface protein data from 500,089 memory T cells to define 31 cell states from 259 individuals in a Peruvian tuberculosis (TB) progression cohort. At immune steady state >4 years after infection and disease resolution, we found that, after accounting for significant effects of age, sex, season and genetic ancestry on T cell composition, a polyfunctional type 17 helper T (TH17) cell-like effector state was reduced in abundance and function in individuals who previously progressed from Mycobacterium tuberculosis (M.tb) infection to active TB disease. These cells are capable of responding to M.tb peptides. Deconvoluting this state-uniquely identifiable with multimodal analysis-from public data demonstrated that its depletion may precede and persist beyond active disease. Our study demonstrates the power of integrative multimodal single-cell profiling to define cell states relevant to disease and other traits.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures















Comment in
-
Mitigating myopia in tuberculosis.Nat Immunol. 2021 Jun;22(6):675-676. doi: 10.1038/s41590-021-00935-z. Nat Immunol. 2021. PMID: 34031615 No abstract available.
Similar articles
-
Mycobacterium tuberculosis multi-drug-resistant strain M induces IL-17+ IFNγ- CD4+ T cell expansion through an IL-23 and TGF-β-dependent mechanism in patients with MDR-TB tuberculosis.Clin Exp Immunol. 2017 Jan;187(1):160-173. doi: 10.1111/cei.12873. Epub 2016 Nov 2. Clin Exp Immunol. 2017. PMID: 27681197 Free PMC article.
-
CD4+ T cell polyfunctional profile in HIV-TB coinfection are similar between individuals with latent and active TB infection.Tuberculosis (Edinb). 2015 Jul;95(4):470-5. doi: 10.1016/j.tube.2014.12.008. Epub 2015 Jan 7. Tuberculosis (Edinb). 2015. PMID: 25956974 Free PMC article.
-
Cell-Mediated Immune Responses to in vivo-Expressed and Stage-Specific Mycobacterium tuberculosis Antigens in Latent and Active Tuberculosis Across Different Age Groups.Front Immunol. 2020 Feb 11;11:103. doi: 10.3389/fimmu.2020.00103. eCollection 2020. Front Immunol. 2020. PMID: 32117257 Free PMC article.
-
IL-22: An Underestimated Player in Natural Resistance to Tuberculosis?Front Immunol. 2018 Sep 25;9:2209. doi: 10.3389/fimmu.2018.02209. eCollection 2018. Front Immunol. 2018. PMID: 30319650 Free PMC article. Review.
-
Th1 and Th17 Cells in Tuberculosis: Protection, Pathology, and Biomarkers.Mediators Inflamm. 2015;2015:854507. doi: 10.1155/2015/854507. Epub 2015 Nov 10. Mediators Inflamm. 2015. PMID: 26640327 Free PMC article. Review.
Cited by
-
Contribution and Future of High-Throughput Transcriptomics in Battling Tuberculosis.Front Microbiol. 2022 Feb 24;13:835620. doi: 10.3389/fmicb.2022.835620. eCollection 2022. Front Microbiol. 2022. PMID: 35283833 Free PMC article. Review.
-
Single cell genomics based insights into the impact of cell-type specific microbial internalization on disease severity.Front Immunol. 2024 May 21;15:1401320. doi: 10.3389/fimmu.2024.1401320. eCollection 2024. Front Immunol. 2024. PMID: 38835769 Free PMC article. Review.
-
A longitudinal single-cell atlas of anti-tumour necrosis factor treatment in inflammatory bowel disease.Nat Immunol. 2024 Nov;25(11):2152-2165. doi: 10.1038/s41590-024-01994-8. Epub 2024 Oct 22. Nat Immunol. 2024. PMID: 39438660 Free PMC article.
-
Characterization and decontamination of background noise in droplet-based single-cell protein expression data with DecontPro.Nucleic Acids Res. 2024 Jan 11;52(1):e4. doi: 10.1093/nar/gkad1032. Nucleic Acids Res. 2024. PMID: 37973397 Free PMC article.
-
Single-cell eQTL models reveal dynamic T cell state dependence of disease loci.Nature. 2022 Jun;606(7912):120-128. doi: 10.1038/s41586-022-04713-1. Epub 2022 May 11. Nature. 2022. PMID: 35545678 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

