Geospatial HIV-1 subtype C gp120 sequence diversity and its predicted impact on broadly neutralizing antibody sensitivity
- PMID: 34029329
- PMCID: PMC8143386
- DOI: 10.1371/journal.pone.0251969
Geospatial HIV-1 subtype C gp120 sequence diversity and its predicted impact on broadly neutralizing antibody sensitivity
Abstract
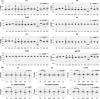
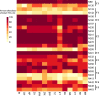
Evolving diversity in globally circulating HIV-1 subtypes presents a formidable challenge in defining and developing neutralizing antibodies for prevention and treatment. HIV-1 subtype C is responsible for majority of global HIV-1 infections. In the present study, we examined the diversity in genetic signatures and attributes that differentiate region-specific HIV-1 subtype C gp120 sequences associated with virus neutralization outcomes to key bnAbs having distinct epitope specificities. A total of 1814 full length HIV-1 subtype C gp120 sequence from 37 countries were retrieved from Los Alamos National Laboratory HIV database (www.hiv.lanl.gov). The amino acid sequences were assessed for their phylogenetic association, variable loop lengths and prevalence of potential N-linked glycosylation sites (pNLGS). Responses of these sequences to bnAbs were predicted with a machine learning algorithm 'bNAb-ReP' and compared with those reported in the CATNAP database. Subtype C sequences from Asian countries including India differed phylogenetically when compared with that from African countries. Variable loop lengths and charges within Indian and African clusters were also found to be distinct from each other, specifically for V1, V2 and V4 loops. Pairwise analyses at each of the 25 pNLG sites indicated distinct country specific profiles. Highly significant differences (p<0.001***) were observed in prevalence of four pNLGS (N130, N295, N392 and N448) between South Africa and India, having most disease burden associated with subtype C. Our findings highlight that distinctly evolving clusters within global intra-subtype C gp120 sequences are likely to influence the disparate region-specific sensitivity of circulating HIV-1 subtype C to bnAbs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Neutralization diversity of HIV-1 Indian subtype C envelopes obtained from cross sectional and followed up individuals against broadly neutralizing monoclonal antibodies having distinct gp120 specificities.Retrovirology. 2021 May 14;18(1):12. doi: 10.1186/s12977-021-00556-2. Retrovirology. 2021. PMID: 33990195 Free PMC article.
-
Subtle Longitudinal Alterations in Env Sequence Potentiate Differences in Sensitivity to Broadly Neutralizing Antibodies following Acute HIV-1 Subtype C Infection.J Virol. 2022 Dec 21;96(24):e0127022. doi: 10.1128/jvi.01270-22. Epub 2022 Dec 1. J Virol. 2022. PMID: 36453881 Free PMC article.
-
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.J Virol. 2016 Jan 13;90(7):3446-57. doi: 10.1128/JVI.03090-15. J Virol. 2016. PMID: 26763999 Free PMC article.
-
HIV-1 subtype C predicted co-receptor tropism in Africa: an individual sequence level meta-analysis.AIDS Res Ther. 2020 Feb 7;17(1):5. doi: 10.1186/s12981-020-0263-x. AIDS Res Ther. 2020. PMID: 32033571 Free PMC article. Review.
-
Antibody responses to the HIV-1 envelope high mannose patch.Adv Immunol. 2019;143:11-73. doi: 10.1016/bs.ai.2019.08.002. Epub 2019 Sep 11. Adv Immunol. 2019. PMID: 31607367 Free PMC article. Review.
Cited by
-
Prevalence of resistance-associated viral variants to the HIV-specific broadly neutralising antibody 10-1074 in a UK bNAb-naïve population.Front Immunol. 2024 Mar 18;15:1352123. doi: 10.3389/fimmu.2024.1352123. eCollection 2024. Front Immunol. 2024. PMID: 38562938 Free PMC article.
-
Web Service for HIV Drug Resistance Prediction Based on Analysis of Amino Acid Substitutions in Main Drug Targets.Viruses. 2023 Nov 11;15(11):2245. doi: 10.3390/v15112245. Viruses. 2023. PMID: 38005921 Free PMC article.
-
Neutralization diversity of HIV-1 Indian subtype C envelopes obtained from cross sectional and followed up individuals against broadly neutralizing monoclonal antibodies having distinct gp120 specificities.Retrovirology. 2021 May 14;18(1):12. doi: 10.1186/s12977-021-00556-2. Retrovirology. 2021. PMID: 33990195 Free PMC article.
-
Subtle Longitudinal Alterations in Env Sequence Potentiate Differences in Sensitivity to Broadly Neutralizing Antibodies following Acute HIV-1 Subtype C Infection.J Virol. 2022 Dec 21;96(24):e0127022. doi: 10.1128/jvi.01270-22. Epub 2022 Dec 1. J Virol. 2022. PMID: 36453881 Free PMC article.
-
Predicted resistance to broadly neutralizing antibodies (bnAbs) and associated HIV-1 envelope characteristics among seroconverting adults in Botswana.Sci Rep. 2023 Oct 24;13(1):18134. doi: 10.1038/s41598-023-44722-2. Sci Rep. 2023. PMID: 37875518 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

