Production of viable chicken by allogeneic transplantation of primordial germ cells induced from somatic cells
- PMID: 34017000
- PMCID: PMC8138025
- DOI: 10.1038/s41467-021-23242-5
Production of viable chicken by allogeneic transplantation of primordial germ cells induced from somatic cells
Retraction in
-
Retraction Note: Production of viable chicken by allogeneic transplantation of primordial germ cells induced from somatic cells.Nat Commun. 2023 Jun 19;14(1):3625. doi: 10.1038/s41467-023-39170-5. Nat Commun. 2023. PMID: 37336948 Free PMC article. No abstract available.
Abstract
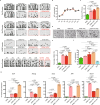
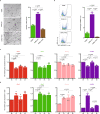
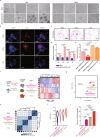
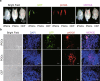
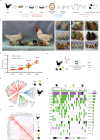
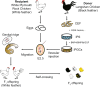
The allogeneic transplantation of primordial germ cells (PGCs) derived from somatic cells overcomes the limitation of avian cloning. Here, we transdifferentiate chicken embryo fibroblasts (CEFs) from black feathered Langshan chickens to PGCs and transplant them into White Plymouth Rock chicken embryos to produce viable offspring with characteristics inherited from the donor. We express Oct4/Sox2/Nanog/Lin28A (OSNL) to reprogram CEFs to induced pluripotent stem cells (iPSCs), which are further induced to differentiate into PGCs by BMP4/BMP8b/EGF. DNA demethylation, histone acetylation and glycolytic activation elevate the iPSC induction efficiency, while histone acetylation and glycolytic inhibition facilitate PGCs formation. The induced PGCs (iPGCs) are transplanted into the recipients, which are self-crossed to produce 189/509 somatic cells derived chicken with the donor's characteristics. Microsatellite analysis and genome sequencing confirm the inheritance of genetic information from the donor. Thus, we demonstrate the feasibility of avian cloning from somatic cells.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Expression dynamics of pluripotency genes in chicken primordial germ cells before and after colonization of the genital ridges.Mol Reprod Dev. 2013 Oct;80(10):849-61. doi: 10.1002/mrd.22216. Epub 2013 Aug 2. Mol Reprod Dev. 2013. PMID: 23877993
-
Direct Reprogramming of Human Primordial Germ Cells into Induced Pluripotent Stem Cells: Efficient Generation of Genetically Engineered Germ Cells.Stem Cells Dev. 2015 Nov 15;24(22):2634-48. doi: 10.1089/scd.2015.0100. Epub 2015 Aug 10. Stem Cells Dev. 2015. PMID: 26154167 Free PMC article.
-
Role of Epigenetic Regulation by the REST/CoREST/HDAC Corepressor Complex of Moderate NANOG Expression in Chicken Primordial Germ Cells.Stem Cells Dev. 2018 Sep 1;27(17):1215-1225. doi: 10.1089/scd.2018.0059. Epub 2018 Aug 20. Stem Cells Dev. 2018. PMID: 30032710
-
Development, differentiation and manipulation of chicken germ cells.Dev Growth Differ. 2013 Jan;55(1):20-40. doi: 10.1111/dgd.12026. Dev Growth Differ. 2013. PMID: 23294359 Review.
-
Pluripotent genes in avian stem cells.Dev Growth Differ. 2013 Jan;55(1):41-51. doi: 10.1111/dgd.12021. Epub 2012 Dec 20. Dev Growth Differ. 2013. PMID: 23278808 Review.
Cited by
-
Dissecting chicken germ cell dynamics by combining a germ cell tracing transgenic chicken model with single-cell RNA sequencing.Comput Struct Biotechnol J. 2022 Apr 2;20:1654-1669. doi: 10.1016/j.csbj.2022.03.040. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35465157 Free PMC article.
-
miR-126 in Extracellular Vesicles Derived from Hepatoblastoma Cells Promotes the Tumorigenesis of Hepatoblastoma through Inducing the Differentiation of BMSCs into Cancer Stem Cells.J Immunol Res. 2021 Oct 29;2021:6744715. doi: 10.1155/2021/6744715. eCollection 2021. J Immunol Res. 2021. PMID: 34746322 Free PMC article.
-
Avian iPSC Derivation to Recover Threatened Wild Species: A Comprehensive Review in Light of Well-Established Protocols.Animals (Basel). 2024 Jan 10;14(2):220. doi: 10.3390/ani14020220. Animals (Basel). 2024. PMID: 38254390 Free PMC article. Review.
-
Poultry genetic heritage cryopreservation and reconstruction: advancement and future challenges.J Anim Sci Biotechnol. 2022 Oct 9;13(1):115. doi: 10.1186/s40104-022-00768-2. J Anim Sci Biotechnol. 2022. PMID: 36210477 Free PMC article. Review.
-
The Transcriptome of Chicken Migratory Primordial Germ Cells Reveals Intrinsic Sex Differences and Expression of Hallmark Germ Cell Genes.Cells. 2023 Apr 13;12(8):1151. doi: 10.3390/cells12081151. Cells. 2023. PMID: 37190060 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

