Cyclophilin A Inhibits Human Respiratory Syncytial Virus (RSV) Replication by Binding to RSV-N through Its PPIase Activity
- PMID: 34011546
- PMCID: PMC8274602
- DOI: 10.1128/JVI.00563-21
Cyclophilin A Inhibits Human Respiratory Syncytial Virus (RSV) Replication by Binding to RSV-N through Its PPIase Activity
Abstract
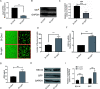
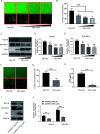
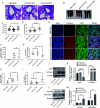
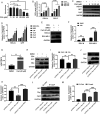
Human respiratory syncytial virus (hRSV) is the most common pathogen which causes acute lower respiratory infection (ALRI) in infants. Recently, virus-host interaction has become a hot spot of virus-related research, and it needs to be further elaborated for RSV infection. In this study, we found that RSV infection significantly increased the expression of cyclophilin A (cypA) in clinical patients, mice, and epithelial cells. Therefore, we evaluated the function of cypA in RSV replication and demonstrated that virus proliferation was accelerated in cypA knockdown host cells but restrained in cypA-overexpressing host cells. Furthermore, we proved that cypA limited RSV replication depending on its PPIase activity. Moreover, we performed liquid chromatography-mass spectrometry, and the results showed that cypA could interact with several viral proteins, such as RSV-N, RSV-P, and RSV-M2-1. Finally, the interaction between cypA and RSV-N was certified by coimmunoprecipitation and immunofluorescence. Those results provided strong evidence that cypA may play an inhibitory role in RSV replication through interaction with RSV-N via its PPIase activity. IMPORTANCE RSV-N, packed in the viral genome to form the ribonucleoprotein (RNP) complex, which is recognized by the RSV RNA-dependent RNA polymerase (RdRp) complex to initiate viral replication and transcription, plays an indispensable role in the viral biosynthesis process. cypA, binding to RSV-N, may impair this function by weakening the interaction between RSV-N and RSV-P, thus leading to decreased viral production. Our research provides novel insight into cypA antiviral function, including binding to viral capsid protein to inhibit viral replication, which may be helpful for new antiviral drug exploration.
Keywords: CSA; PPIase; RSV-N; cypA; viral replication.
Figures



Similar articles
-
Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells.J Virol. 2005 Jan;79(1):176-83. doi: 10.1128/JVI.79.1.176-183.2005. J Virol. 2005. PMID: 15596813 Free PMC article.
-
Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex.J Virol. 2009 Jul;83(13):6554-65. doi: 10.1128/JVI.02550-08. Epub 2009 Apr 22. J Virol. 2009. PMID: 19386705 Free PMC article.
-
Respiratory Syncytial Virus-Induced Oxidative Stress Leads to an Increase in Labile Zinc Pools in Lung Epithelial Cells.mSphere. 2020 May 27;5(3):e00447-20. doi: 10.1128/mSphere.00447-20. mSphere. 2020. PMID: 32461278 Free PMC article.
-
Cyclophilin A: A Key Factor in Virus Replication and Potential Target for Anti-viral Therapy.Curr Issues Mol Biol. 2017;21:1-20. doi: 10.21775/cimb.021.001. Epub 2016 Mar 31. Curr Issues Mol Biol. 2017. PMID: 27033630 Review.
-
Insights into the roles of cyclophilin A during influenza virus infection.Viruses. 2013 Jan 15;5(1):182-91. doi: 10.3390/v5010182. Viruses. 2013. PMID: 23322171 Free PMC article. Review.
Cited by
-
Cyclophilin A facilitates influenza B virus replication by stabilizing viral proteins.iScience. 2023 Nov 23;26(12):108515. doi: 10.1016/j.isci.2023.108515. eCollection 2023 Dec 15. iScience. 2023. PMID: 38089580 Free PMC article.
-
Respiratory syncytial virus infection and novel interventions.Nat Rev Microbiol. 2023 Nov;21(11):734-749. doi: 10.1038/s41579-023-00919-w. Epub 2023 Jul 12. Nat Rev Microbiol. 2023. PMID: 37438492 Review.
-
The effect of overexpression of CyPA on gene expression in human umbilical vein endothelial cells.Medicine (Baltimore). 2024 Jul 19;103(29):e38886. doi: 10.1097/MD.0000000000038886. Medicine (Baltimore). 2024. PMID: 39029007 Free PMC article.
References
-
- Pneumonia Etiology Research for Child Health (PERCH) Study Group. 2019. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet 394:757–779. 10.1016/S0140-6736(19)30721-4. [Erratum, 394: 736, 2019, 10.1016/S0140-6736(19)32010-0.] - DOI - DOI - PMC - PubMed
-
- Chaw PS, Hua L, Cunningham S, Campbell H, Mikolajczyk R, Nair H, RESCEU Investigators . 2020. Respiratory syncytial virus-associated acute lower respiratory infections in children with bronchopulmonary dysplasia: systematic review and meta-analysis. J Infect Dis 222(Suppl 7):S620–S627. 10.1093/infdis/jiz492. - DOI - PubMed
-
- Chaw PS, Wong SWL, Cunningham S, Campbell H, Mikolajczyk R, Nair H, RESCEU Investigators . 2020. Acute lower respiratory infections associated with respiratory syncytial virus in children with underlying congenital heart disease: systematic review and meta-analysis. J Infect Dis 222(Suppl 7):S613–S619. 10.1093/infdis/jiz150. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

