Regeneration linked miRNA modify tumor phenotype and can enforce multi-lineage growth arrest in vivo
- PMID: 34006907
- PMCID: PMC8131690
- DOI: 10.1038/s41598-021-90009-9
Regeneration linked miRNA modify tumor phenotype and can enforce multi-lineage growth arrest in vivo
Abstract
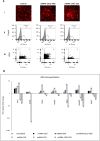
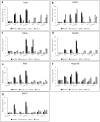
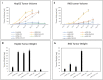
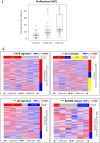
Regulated cell proliferation is an effector mechanism of regeneration, whilst dysregulated cell proliferation is a feature of cancer. We have previously identified microRNA (miRNA) that regulate successful and failed human liver regeneration. We hypothesized that these regulators may directly modify tumor behavior. Here we show that inhibition of miRNAs -503 and -23a, alone or in combination, enhances tumor proliferation in hepatocyte and non-hepatocyte derived cancers in vitro, driving more aggressive tumor behavior in vivo. Inhibition of miRNA-152 caused induction of DNMT1, site-specific methylation with associated changes in gene expression and in vitro and in vivo growth inhibition. Enforced changes in expression of two miRNA recapitulating changes observed in failed regeneration led to complete growth inhibition of multi-lineage cancers in vivo. Our results indicate that regulation of regeneration and tumor aggressiveness are concordant and that miRNA-based inhibitors of regeneration may constitute a novel treatment strategy for human cancers.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Inhibition of miR-21 rescues liver regeneration after partial hepatectomy in ethanol-fed rats.Am J Physiol Gastrointest Liver Physiol. 2016 Nov 1;311(5):G794-G806. doi: 10.1152/ajpgi.00292.2016. Epub 2016 Sep 15. Am J Physiol Gastrointest Liver Physiol. 2016. PMID: 27634014 Free PMC article.
-
Role of microRNA in liver regeneration.Hepatobiliary Pancreat Dis Int. 2016 Apr;15(2):141-6. doi: 10.1016/s1499-3872(15)60036-4. Hepatobiliary Pancreat Dis Int. 2016. PMID: 27020629 Review.
-
MicroRNAs control hepatocyte proliferation during liver regeneration.Hepatology. 2010 May;51(5):1735-43. doi: 10.1002/hep.23547. Hepatology. 2010. PMID: 20432256 Free PMC article.
-
Genomewide microRNA down-regulation as a negative feedback mechanism in the early phases of liver regeneration.Hepatology. 2011 Aug;54(2):609-19. doi: 10.1002/hep.24421. Epub 2011 Jun 30. Hepatology. 2011. PMID: 21574170 Free PMC article.
-
The role of microRNAs in liver injury at the crossroad between hepatic cell death and regeneration.Biochem Biophys Res Commun. 2017 Jan 15;482(3):399-407. doi: 10.1016/j.bbrc.2016.10.084. Epub 2016 Oct 24. Biochem Biophys Res Commun. 2017. PMID: 27789285 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

