Post-translational Control of RNA-Binding Proteins and Disease-Related Dysregulation
- PMID: 33987205
- PMCID: PMC8111222
- DOI: 10.3389/fmolb.2021.658852
Post-translational Control of RNA-Binding Proteins and Disease-Related Dysregulation
Abstract
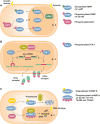
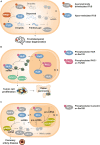
Cell signaling mechanisms modulate gene expression in response to internal and external stimuli. Cellular adaptation requires a precise and coordinated regulation of the transcription and translation processes. The post-transcriptional control of mRNA metabolism is mediated by the so-called RNA-binding proteins (RBPs), which assemble with specific transcripts forming messenger ribonucleoprotein particles of highly dynamic composition. RBPs constitute a class of trans-acting regulatory proteins with affinity for certain consensus elements present in mRNA molecules. However, these regulators are subjected to post-translational modifications (PTMs) that constantly adjust their activity to maintain cell homeostasis. PTMs can dramatically change the subcellular localization, the binding affinity for RNA and protein partners, and the turnover rate of RBPs. Moreover, the ability of many RBPs to undergo phase transition and/or their recruitment to previously formed membrane-less organelles, such as stress granules, is also regulated by specific PTMs. Interestingly, the dysregulation of PTMs in RBPs has been associated with the pathophysiology of many different diseases. Abnormal PTM patterns can lead to the distortion of the physiological role of RBPs due to mislocalization, loss or gain of function, and/or accelerated or disrupted degradation. This Mini Review offers a broad overview of the post-translational regulation of selected RBPs and the involvement of their dysregulation in neurodegenerative disorders, cancer and other pathologies.
Keywords: FUS; HuR; KSRP; RNA-binding proteins; TIA-1/TIAR; hnRNP K; liquid–liquid phase separation; post-translational modifications.
Copyright © 2021 Velázquez-Cruz, Baños-Jaime, Díaz-Quintana, De la Rosa and Díaz-Moreno.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics.J Biol Chem. 2019 May 3;294(18):7137-7150. doi: 10.1074/jbc.TM118.001189. Epub 2018 Dec 26. J Biol Chem. 2019. PMID: 30587571 Free PMC article. Review.
-
Fates and functions of RNA-binding proteins under stress.Wiley Interdiscip Rev RNA. 2023 Nov 28:e1825. doi: 10.1002/wrna.1825. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 38014833 Review.
-
RNA Binding Protein Regulation and Cross-Talk in the Control of AU-rich mRNA Fate.Front Mol Biosci. 2017 Oct 23;4:71. doi: 10.3389/fmolb.2017.00071. eCollection 2017. Front Mol Biosci. 2017. PMID: 29109951 Free PMC article. Review.
-
Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs.Mol Cell Biol. 2007 Sep;27(18):6265-78. doi: 10.1128/MCB.00500-07. Epub 2007 Jul 9. Mol Cell Biol. 2007. PMID: 17620417 Free PMC article.
-
AU-Rich Element RNA Binding Proteins: At the Crossroads of Post-Transcriptional Regulation and Genome Integrity.Int J Mol Sci. 2021 Dec 22;23(1):96. doi: 10.3390/ijms23010096. Int J Mol Sci. 2021. PMID: 35008519 Free PMC article. Review.
Cited by
-
RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation.Viruses. 2022 Jan 19;14(2):188. doi: 10.3390/v14020188. Viruses. 2022. PMID: 35215780 Free PMC article. Review.
-
An AKT1-and TRIM21-mediated phosphodegron controls proteasomal degradation of HuR enabling cell survival under heat shock.iScience. 2023 Feb 28;26(4):106307. doi: 10.1016/j.isci.2023.106307. eCollection 2023 Apr 21. iScience. 2023. PMID: 36968077 Free PMC article.
-
Tankyrase-1 regulates RBP-mediated mRNA turnover to promote muscle fiber formation.Nucleic Acids Res. 2024 Apr 24;52(7):4002-4020. doi: 10.1093/nar/gkae059. Nucleic Acids Res. 2024. PMID: 38321934 Free PMC article.
-
Posttranscriptional Regulation of the Plasminogen Activation System by Non-Coding RNA in Cancer.Int J Mol Sci. 2023 Jan 4;24(2):962. doi: 10.3390/ijms24020962. Int J Mol Sci. 2023. PMID: 36674481 Free PMC article. Review.
-
ADP-Ribosylation Post-Translational Modification: An Overview with a Focus on RNA Biology and New Pharmacological Perspectives.Biomolecules. 2022 Mar 13;12(3):443. doi: 10.3390/biom12030443. Biomolecules. 2022. PMID: 35327636 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

