An Immune Checkpoint-Related Gene Signature for Predicting Survival of Pediatric Acute Myeloid Leukemia
- PMID: 33986802
- PMCID: PMC8079183
- DOI: 10.1155/2021/5550116
An Immune Checkpoint-Related Gene Signature for Predicting Survival of Pediatric Acute Myeloid Leukemia
Abstract
Objective: The aim of this research was to create a new genetic signature of immune checkpoint-associated genes as a prognostic method for pediatric acute myeloid leukemia (AML).
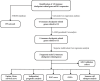
Methods: Transcriptome profiles and clinical follow-up details were obtained in Therapeutically Applicable Research to Generate Effective Treatments (TARGET), a database of pediatric tumors. Secondary data was collected from the Gene Expression Omnibus (GEO) to test the observations. In univariate Cox regression and multivariate Cox regression studies, the expression of immune checkpoint-related genes was studied. A three-mRNA signature was developed for predicting pediatric AML patient survival. Furthermore, the GEO cohort was used to confirm the reliability. A bioinformatics method was utilized to identify the diagnostic and prognostic value.
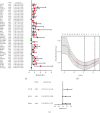
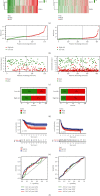
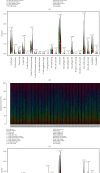
Results: A three-gene (STAT1, BATF, EML4) signature was developed to identify patients into two danger categories depending on their OS. A multivariate regression study showed that the immune checkpoint-related signature (STAT1, BATF, EML4) was an independent indicator of pediatric AML. By immune cell subtypes analyses, the signature was correlated with multiple subtypes of immune cells.
Conclusion: In summary, our three-gene signature can be a useful tool to predict the OS in AML patients.
Copyright © 2021 Feng Jiang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

Similar articles
-
Prognosis and Characterization of Immune Microenvironment in Acute Myeloid Leukemia Through Identification of an Autophagy-Related Signature.Front Immunol. 2021 May 31;12:695865. doi: 10.3389/fimmu.2021.695865. eCollection 2021. Front Immunol. 2021. PMID: 34135913 Free PMC article.
-
Construction of a Pyroptosis-Related Signature for Prognostic Prediction and Characterization of Immune Microenvironment in Acute Myelogenous Leukemia.Int J Gen Med. 2022 Mar 12;15:2913-2927. doi: 10.2147/IJGM.S352062. eCollection 2022. Int J Gen Med. 2022. PMID: 35308573 Free PMC article.
-
Identification of a Mitochondria-Related Gene Signature to Predict the Prognosis in AML.Front Oncol. 2022 Mar 10;12:823831. doi: 10.3389/fonc.2022.823831. eCollection 2022. Front Oncol. 2022. PMID: 35359394 Free PMC article.
-
Identification of an Immune-Related Gene Signature Based on Immunogenomic Landscape Analysis to Predict the Prognosis of Adult Acute Myeloid Leukemia Patients.Front Oncol. 2020 Nov 20;10:574939. doi: 10.3389/fonc.2020.574939. eCollection 2020. Front Oncol. 2020. PMID: 33330048 Free PMC article.
-
Development and External Validation of a Novel Immune Checkpoint-Related Gene Signature for Prediction of Overall Survival in Hepatocellular Carcinoma.Front Mol Biosci. 2021 Jan 21;7:620765. doi: 10.3389/fmolb.2020.620765. eCollection 2020. Front Mol Biosci. 2021. PMID: 33553243 Free PMC article.
Cited by
-
Effects of Combinatory In Vitro Treatment with Immune Checkpoint Inhibitors and Cytarabine on the Anti-Cancer Immune Microenvironment in De Novo AML Patients.Cancers (Basel). 2024 Jan 22;16(2):462. doi: 10.3390/cancers16020462. Cancers (Basel). 2024. PMID: 38275902 Free PMC article.
-
Exploring Prognostic Biomarkers of Acute Myeloid Leukemia to Determine Its Most Effective Drugs from the FDA-Approved List through Molecular Docking and Dynamic Simulation.Biomed Res Int. 2023 Jun 15;2023:1946703. doi: 10.1155/2023/1946703. eCollection 2023. Biomed Res Int. 2023. PMID: 37359050 Free PMC article.
-
Integrated analysis of single-cell RNA-seq and bulk RNA-seq reveals RNA N6-methyladenosine modification associated with prognosis and drug resistance in acute myeloid leukemia.Front Immunol. 2023 Oct 31;14:1281687. doi: 10.3389/fimmu.2023.1281687. eCollection 2023. Front Immunol. 2023. PMID: 38022588 Free PMC article.
-
Comprehensive analysis for cellular senescence-related immunogenic characteristics and immunotherapy prediction of acute myeloid leukemia.Front Pharmacol. 2022 Sep 26;13:987398. doi: 10.3389/fphar.2022.987398. eCollection 2022. Front Pharmacol. 2022. PMID: 36225590 Free PMC article.
-
Construction and validation model of necroptosis-related gene signature associates with immunity for osteosarcoma patients.Sci Rep. 2022 Sep 23;12(1):15893. doi: 10.1038/s41598-022-20217-4. Sci Rep. 2022. PMID: 36151259 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

