Unlocking the PIP-box: A peptide library reveals interactions that drive high-affinity binding to human PCNA
- PMID: 33984330
- PMCID: PMC8191301
- DOI: 10.1016/j.jbc.2021.100773
Unlocking the PIP-box: A peptide library reveals interactions that drive high-affinity binding to human PCNA
Abstract
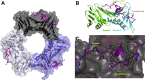
The human sliding clamp, Proliferating Cell Nuclear Antigen (hPCNA), interacts with over 200 proteins through a conserved binding motif, the PIP-box, to orchestrate DNA replication and repair. It is not clear how changes to the features of a PIP-box modulate protein binding and thus how they fine-tune downstream processes. Here, we present a systematic study of each position within the PIP-box to reveal how hPCNA-interacting peptides bind with drastically varied affinities. We synthesized a series of 27 peptides derived from the native protein p21 with small PIP-box modifications and another series of 19 peptides containing PIP-box binding motifs from other proteins. The hPCNA-binding affinity of all peptides, characterized as KD values determined by surface plasmon resonance, spanned a 4000-fold range, from 1.83 nM to 7.59 μM. The hPCNA-bound peptide structures determined by X-ray crystallography and modeled computationally revealed intermolecular and intramolecular interaction networks that correlate with high hPCNA affinity. These data informed rational design of three new PIP-box sequences, testing of which revealed the highest affinity hPCNA-binding partner to date, with a KD value of 1.12 nM, from a peptide with PIP-box QTRITEYF. This work showcases the sequence-specific nuances within the PIP-box that are responsible for high-affinity hPCNA binding, which underpins our understanding of how nature tunes hPCNA affinity to regulate DNA replication and repair processes. In addition, these insights will be useful to future design of hPCNA inhibitors.
Keywords: crystallography; peptides; proliferating cell nuclear antigen (PCNA); structural biology; surface plasmon resonance (SPR).
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

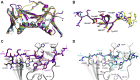


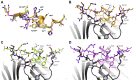
Similar articles
-
Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1.Structure. 2004 Dec;12(12):2209-19. doi: 10.1016/j.str.2004.09.018. Structure. 2004. PMID: 15576034
-
Rational Design of a 310 -Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.Chemistry. 2018 Aug 6;24(44):11325-11331. doi: 10.1002/chem.201801734. Epub 2018 Jul 19. Chemistry. 2018. PMID: 29917264
-
Structure of the sliding clamp from the fungal pathogen Aspergillus fumigatus (AfumPCNA) and interactions with Human p21.FEBS J. 2017 Mar;284(6):985-1002. doi: 10.1111/febs.14035. Epub 2017 Feb 27. FEBS J. 2017. PMID: 28165677
-
Targeting PCNA with Peptide Mimetics for Therapeutic Purposes.Chembiochem. 2020 Feb 17;21(4):442-450. doi: 10.1002/cbic.201900275. Epub 2019 Oct 15. Chembiochem. 2020. PMID: 31247123 Review.
-
Human PCNA Structure, Function and Interactions.Biomolecules. 2020 Apr 8;10(4):570. doi: 10.3390/biom10040570. Biomolecules. 2020. PMID: 32276417 Free PMC article. Review.
Cited by
-
X-ray crystal structure of proliferating cell nuclear antigen 1 from Aeropyrum pernix.Acta Crystallogr F Struct Biol Commun. 2024 Nov 1;80(Pt 11):294-301. doi: 10.1107/S2053230X24009518. Epub 2024 Oct 9. Acta Crystallogr F Struct Biol Commun. 2024. PMID: 39382846 Free PMC article.
-
Structure of the PCNA unloader Elg1-RFC.Sci Adv. 2024 Mar;10(9):eadl1739. doi: 10.1126/sciadv.adl1739. Epub 2024 Mar 1. Sci Adv. 2024. PMID: 38427736 Free PMC article.
-
D-type cyclins regulate DNA mismatch repair in the G1 and S phases of the cell cycle, maintaining genome stability.bioRxiv [Preprint]. 2024 Jan 13:2024.01.12.575420. doi: 10.1101/2024.01.12.575420. bioRxiv. 2024. Update in: Mol Cell. 2024 Apr 4;84(7):1224-1242.e13. doi: 10.1016/j.molcel.2024.02.010 PMID: 38260436 Free PMC article. Updated. Preprint.
-
Towards a High-Affinity Peptidomimetic Targeting Proliferating Cell Nuclear Antigen from Aspergillus fumigatus.J Fungi (Basel). 2023 Nov 10;9(11):1098. doi: 10.3390/jof9111098. J Fungi (Basel). 2023. PMID: 37998903 Free PMC article.
-
Autophosphorylation of the Tousled-like kinases TLK1 and TLK2 regulates recruitment to damaged chromatin via PCNA interaction.bioRxiv [Preprint]. 2024 Apr 26:2024.04.22.590659. doi: 10.1101/2024.04.22.590659. bioRxiv. 2024. Update in: Nucleic Acids Res. 2024 Dec 27:gkae1279. doi: 10.1093/nar/gkae1279 PMID: 38712247 Free PMC article. Updated. Preprint.
References
-
- De Biasio A., Blanco F.J. Proliferating cell nuclear antigen structure and interactions: Too many partners for one dancer? Adv. Protein Chem. Struct. Biol. 2013;91:1–36. - PubMed
-
- Tsurimoto T. PCNA binding proteins. Front. Biosci. 1999;4:D849–D858. - PubMed
-
- Maga G., Hubscher U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 2003;116:3051–3060. - PubMed
-
- Moldovan G.L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

