S-acylation of P2K1 mediates extracellular ATP-induced immune signaling in Arabidopsis
- PMID: 33980819
- PMCID: PMC8115640
- DOI: 10.1038/s41467-021-22854-1
S-acylation of P2K1 mediates extracellular ATP-induced immune signaling in Arabidopsis
Abstract
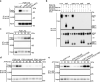
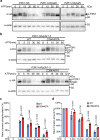
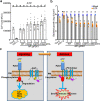
S-acylation is a reversible protein post-translational modification mediated by protein S-acyltransferases (PATs). How S-acylation regulates plant innate immunity is our main concern. Here, we show that the plant immune receptor P2K1 (DORN1, LecRK-I.9; extracellular ATP receptor) directly interacts with and phosphorylates Arabidopsis PAT5 and PAT9 to stimulate their S-acyltransferase activity. This leads, in a time-dependent manner, to greater S-acylation of P2K1, which dampens the immune response. pat5 and pat9 mutants have an elevated extracellular ATP-induced immune response, limited bacterial invasion, increased phosphorylation and decreased degradation of P2K1 during immune signaling. Mutation of S-acylated cysteine residues in P2K1 results in a similar phenotype. Our study reveals that S-acylation effects the temporal dynamics of P2K1 receptor activity, through autophosphorylation and protein degradation, suggesting an important role for this modification in regulating the ability of plants in respond to external stimuli.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Phosphorylation-mediated regulation of integrin-linked kinase 5 by purinoreceptor P2K2.Plant Signal Behav. 2023 Dec 31;18(1):2261743. doi: 10.1080/15592324.2023.2261743. Epub 2023 Dec 17. Plant Signal Behav. 2023. PMID: 37750411 Free PMC article.
-
Arabidopsis Lectin Receptor Kinase P2K2 Is a Second Plant Receptor for Extracellular ATP and Contributes to Innate Immunity.Plant Physiol. 2020 Jul;183(3):1364-1375. doi: 10.1104/pp.19.01265. Epub 2020 Apr 28. Plant Physiol. 2020. PMID: 32345768 Free PMC article.
-
Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity.Proc Natl Acad Sci U S A. 2017 May 30;114(22):5749-5754. doi: 10.1073/pnas.1614468114. Epub 2017 May 15. Proc Natl Acad Sci U S A. 2017. PMID: 28507137 Free PMC article.
-
Molecular Mechanism of Plant Recognition of Extracellular ATP.Adv Exp Med Biol. 2017;1051:233-253. doi: 10.1007/5584_2017_110. Adv Exp Med Biol. 2017. PMID: 29064066 Review.
-
Extracellular ATP, a danger signal, is recognized by DORN1 in Arabidopsis.Biochem J. 2014 Nov 1;463(3):429-37. doi: 10.1042/BJ20140666. Biochem J. 2014. PMID: 25301072 Review.
Cited by
-
Arabidopsis protein S-acyl transferases positively mediate BR signaling through S-acylation of BSK1.Proc Natl Acad Sci U S A. 2024 Feb 13;121(7):e2322375121. doi: 10.1073/pnas.2322375121. Epub 2024 Feb 5. Proc Natl Acad Sci U S A. 2024. PMID: 38315835 Free PMC article.
-
Phosphorylation-mediated regulation of integrin-linked kinase 5 by purinoreceptor P2K2.Plant Signal Behav. 2023 Dec 31;18(1):2261743. doi: 10.1080/15592324.2023.2261743. Epub 2023 Dec 17. Plant Signal Behav. 2023. PMID: 37750411 Free PMC article.
-
Unveiling orphan receptor-like kinases in plants: novel client discovery using high-confidence library predictions in the Kinase-Client (KiC) assay.Front Plant Sci. 2024 Apr 3;15:1372361. doi: 10.3389/fpls.2024.1372361. eCollection 2024. Front Plant Sci. 2024. PMID: 38633461 Free PMC article.
-
The S-acylation cycle of transcription factor MtNAC80 influences cold stress responses in Medicago truncatula.Plant Cell. 2024 Jul 2;36(7):2629-2651. doi: 10.1093/plcell/koae103. Plant Cell. 2024. PMID: 38552172
-
The intrinsically disordered C-terminus of purinoceptor P2K1 fine-tunes plant responses to extracellular ATP.FEBS Lett. 2023 Aug;597(16):2059-2071. doi: 10.1002/1873-3468.14703. Epub 2023 Jul 27. FEBS Lett. 2023. PMID: 37465901 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

