Smc5/6 in the rDNA modulates lifespan independently of Fob1
- PMID: 33979898
- PMCID: PMC8208791
- DOI: 10.1111/acel.13373
Smc5/6 in the rDNA modulates lifespan independently of Fob1
Abstract
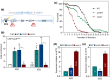
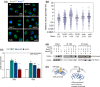
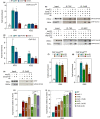
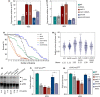
The ribosomal DNA (rDNA) in Saccharomyces cerevisiae is in one tandem repeat array on Chromosome XII. Two regions within each repetitive element, called intergenic spacer 1 (IGS1) and IGS2, are important for organizing the rDNA within the nucleolus. The Smc5/6 complex localizes to IGS1 and IGS2. We show that Smc5/6 has a function in the rDNA beyond its role in homologous recombination (HR) at the replication fork barrier (RFB) located in IGS1. Fob1 is required for optimal binding of Smc5/6 at IGS1 whereas the canonical silencing factor Sir2 is required for its optimal binding at IGS2, independently of Fob1. Through interdependent interactions, Smc5/6 stabilizes Sir2 and Cohibin at both IGS and its recovery at IGS2 is important for nucleolar compaction and transcriptional silencing, which in turn supports rDNA stability and lifespan.
Keywords: Fob1; Smc5/6; nucleolar morphology; nucleolus; rDNA; replicative lifespan; silencing.
© 2021 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
Replication fork arrest and rDNA silencing are two independent and separable functions of the replication terminator protein Fob1 of Saccharomyces cerevisiae.J Biol Chem. 2010 Apr 23;285(17):12612-9. doi: 10.1074/jbc.M109.082388. Epub 2010 Feb 23. J Biol Chem. 2010. PMID: 20179323 Free PMC article.
-
Acute Smc5/6 depletion reveals its primary role in rDNA replication by restraining recombination at fork pausing sites.PLoS Genet. 2018 Jan 23;14(1):e1007129. doi: 10.1371/journal.pgen.1007129. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29360860 Free PMC article.
-
Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing.Genes Dev. 2003 Sep 1;17(17):2162-76. doi: 10.1101/gad.1108403. Epub 2003 Aug 15. Genes Dev. 2003. PMID: 12923057 Free PMC article.
-
Saccharomyces cerevisiae rDNA as super-hub: the region where replication, transcription and recombination meet.Cell Mol Life Sci. 2020 Dec;77(23):4787-4798. doi: 10.1007/s00018-020-03562-3. Epub 2020 May 31. Cell Mol Life Sci. 2020. PMID: 32476055 Free PMC article. Review.
-
Regulation of rDNA stability by sumoylation.DNA Repair (Amst). 2009 Apr 5;8(4):507-16. doi: 10.1016/j.dnarep.2009.01.015. Epub 2009 Mar 3. DNA Repair (Amst). 2009. PMID: 19261548 Review.
Cited by
-
The multi-functional Smc5/6 complex in genome protection and disease.Nat Struct Mol Biol. 2023 Jun;30(6):724-734. doi: 10.1038/s41594-023-01015-6. Epub 2023 Jun 19. Nat Struct Mol Biol. 2023. PMID: 37336994 Free PMC article. Review.
-
Nucleolar release of rDNA repeats for repair involves SUMO-mediated untethering by the Cdc48/p97 segregase.Nat Commun. 2021 Aug 13;12(1):4918. doi: 10.1038/s41467-021-25205-2. Nat Commun. 2021. PMID: 34389719 Free PMC article.
-
SIR telomere silencing depends on nuclear envelope lipids and modulates sensitivity to a lysolipid.J Cell Biol. 2023 Jul 3;222(7):e202206061. doi: 10.1083/jcb.202206061. Epub 2023 Apr 12. J Cell Biol. 2023. PMID: 37042812 Free PMC article.
-
The SMC5/6 complex: folding chromosomes back into shape when genomes take a break.Nucleic Acids Res. 2024 Mar 21;52(5):2112-2129. doi: 10.1093/nar/gkae103. Nucleic Acids Res. 2024. PMID: 38375830 Free PMC article. Review.
References
-
- Brewer, B. J. , & Fangman, W. L. (1988). A replication fork barrier at the 3’ end of yeast ribosomal RNA genes. Cell, 55, 637–643. - PubMed
-
- Bryk, M. , Banerjee, M. , Murphy, M. , Knudsen, K. E. , Garfinkel, D. J. , & Curcio, M. J. (1997). Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Development, 11, 255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

