Methylation of histone H3 at lysine 37 by Set1 and Set2 prevents spurious DNA replication
- PMID: 33979575
- PMCID: PMC7612968
- DOI: 10.1016/j.molcel.2021.04.021
Methylation of histone H3 at lysine 37 by Set1 and Set2 prevents spurious DNA replication
Abstract
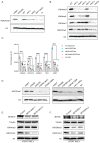
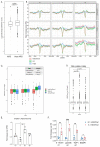
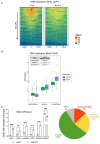
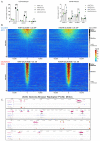
DNA replication initiates at genomic locations known as origins of replication, which, in S. cerevisiae, share a common DNA consensus motif. Despite being virtually nucleosome-free, origins of replication are greatly influenced by the surrounding chromatin state. Here, we show that histone H3 lysine 37 mono-methylation (H3K37me1) is catalyzed by Set1p and Set2p and that it regulates replication origin licensing. H3K37me1 is uniformly distributed throughout most of the genome, but it is scarce at replication origins, where it increases according to the timing of their firing. We find that H3K37me1 hinders Mcm2 interaction with chromatin, maintaining low levels of MCM outside of conventional replication origins. Lack of H3K37me1 results in defective DNA replication from canonical origins while promoting replication events at inefficient and non-canonical sites. Collectively, our results indicate that H3K37me1 ensures correct execution of the DNA replication program by protecting the genome from inappropriate origin licensing and spurious DNA replication.
Keywords: H3K37methylation; Histone modifications; MCM; Origin licensing; Replication origins; Set1; Set2.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.K. is a co-founder and shareholder of Abcam Plc and a co-founder of Storm Therapeutics Ltd. (Cambridge, UK). T.L. is a consultant for Storm Therapeutics Ltd.
Figures


Similar articles
-
Diverse and dynamic forms of gene regulation by the S. cerevisiae histone methyltransferase Set1.Curr Genet. 2023 Jun;69(2-3):91-114. doi: 10.1007/s00294-023-01265-3. Epub 2023 Mar 31. Curr Genet. 2023. PMID: 37000206 Review.
-
H3 k36 methylation helps determine the timing of cdc45 association with replication origins.PLoS One. 2009 Jun 12;4(6):e5882. doi: 10.1371/journal.pone.0005882. PLoS One. 2009. PMID: 19521516 Free PMC article.
-
RNA Binding by Histone Methyltransferases Set1 and Set2.Mol Cell Biol. 2017 Jun 29;37(14):e00165-17. doi: 10.1128/MCB.00165-17. Print 2017 Jul 15. Mol Cell Biol. 2017. PMID: 28483910 Free PMC article.
-
Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe.Eukaryot Cell. 2005 Aug;4(8):1446-54. doi: 10.1128/EC.4.8.1446-1454.2005. Eukaryot Cell. 2005. PMID: 16087749 Free PMC article.
-
Structural basis for H3K4 trimethylation by yeast Set1/COMPASS.Adv Enzyme Regul. 2010;50(1):104-10. doi: 10.1016/j.advenzreg.2009.12.005. Epub 2009 Dec 18. Adv Enzyme Regul. 2010. PMID: 20005892 Free PMC article. Review.
Cited by
-
The protein methylation network in yeast: A landmark in completeness for a eukaryotic post-translational modification.Proc Natl Acad Sci U S A. 2023 Jun 6;120(23):e2215431120. doi: 10.1073/pnas.2215431120. Epub 2023 May 30. Proc Natl Acad Sci U S A. 2023. PMID: 37252976 Free PMC article.
-
SETD2: from chromatin modifier to multipronged regulator of the genome and beyond.Cell Mol Life Sci. 2022 Jun 6;79(6):346. doi: 10.1007/s00018-022-04352-9. Cell Mol Life Sci. 2022. PMID: 35661267 Free PMC article. Review.
-
SMYD5 is a histone H3-specific methyltransferase mediating mono-methylation of histone H3 lysine 36 and 37.Biochem Biophys Res Commun. 2022 Apr 9;599:142-147. doi: 10.1016/j.bbrc.2022.02.043. Epub 2022 Feb 12. Biochem Biophys Res Commun. 2022. PMID: 35182940 Free PMC article.
-
Where and when to start: Regulating DNA replication origin activity in eukaryotic genomes.Nucleus. 2023 Dec;14(1):2229642. doi: 10.1080/19491034.2023.2229642. Nucleus. 2023. PMID: 37469113 Free PMC article. Review.
-
Diverse and dynamic forms of gene regulation by the S. cerevisiae histone methyltransferase Set1.Curr Genet. 2023 Jun;69(2-3):91-114. doi: 10.1007/s00294-023-01265-3. Epub 2023 Mar 31. Curr Genet. 2023. PMID: 37000206 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- WT203144/WT_/Wellcome Trust/United Kingdom
- RG72100/CRUK_/Cancer Research UK/United Kingdom
- 27321/CRUK_/Cancer Research UK/United Kingdom
- 210926/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- A18061/CRUK_/Cancer Research UK/United Kingdom
- RG96894/CRUK_/Cancer Research UK/United Kingdom
- 17001/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 309952/ERC_/European Research Council/International
- RG94424/WT_/Wellcome Trust/United Kingdom
- C6946/A24843/CRUK_/Cancer Research UK/United Kingdom
- 210926/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

