Synthetic Biology: Emerging Concepts to Design and Advance Adeno-Associated Viral Vectors for Gene Therapy
- PMID: 33977059
- PMCID: PMC8097373
- DOI: 10.1002/advs.202004018
Synthetic Biology: Emerging Concepts to Design and Advance Adeno-Associated Viral Vectors for Gene Therapy
Abstract
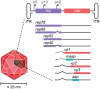
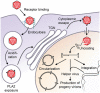
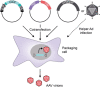
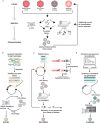
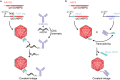
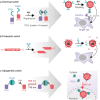
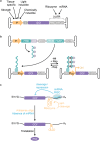
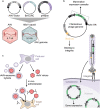
Three recent approvals and over 100 ongoing clinical trials make adeno-associated virus (AAV)-based vectors the leading gene delivery vehicles in gene therapy. Pharmaceutical companies are investing in this small and nonpathogenic gene shuttle to increase the therapeutic portfolios within the coming years. This prospect of marking a new era in gene therapy has fostered both investigations of the fundamental AAV biology as well as engineering studies to enhance delivery vehicles. Driven by the high clinical potential, a new generation of synthetic-biologically engineered AAV vectors is on the rise. Concepts from synthetic biology enable the control and fine-tuning of vector function at different stages of cellular transduction and gene expression. It is anticipated that the emerging field of synthetic-biologically engineered AAV vectors can shape future gene therapeutic approaches and thus the design of tomorrow's gene delivery vectors. This review describes and discusses the recent trends in capsid and vector genome engineering, with particular emphasis on synthetic-biological approaches.
Keywords: AAV; adeno‐associated virus; capsid modifications; engineering; gene delivery; molecular switches; vector design.
© 2021 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Small But Increasingly Mighty: Latest Advances in AAV Vector Research, Design, and Evolution.Hum Gene Ther. 2017 Nov;28(11):1075-1086. doi: 10.1089/hum.2017.172. Epub 2017 Aug 23. Hum Gene Ther. 2017. PMID: 28835125 Review.
-
Adeno-associated virus (AAV) vectors in cancer gene therapy.J Control Release. 2016 Oct 28;240:287-301. doi: 10.1016/j.jconrel.2016.01.001. Epub 2016 Jan 12. J Control Release. 2016. PMID: 26796040 Free PMC article. Review.
-
Engineering and evolution of synthetic adeno-associated virus (AAV) gene therapy vectors via DNA family shuffling.J Vis Exp. 2012 Apr 2;(62):3819. doi: 10.3791/3819. J Vis Exp. 2012. PMID: 22491297 Free PMC article.
-
Generation of Targeted Adeno-Associated Virus (AAV) Vectors for Human Gene Therapy.Curr Pharm Des. 2015;21(22):3248-56. doi: 10.2174/1381612821666150531171653. Curr Pharm Des. 2015. PMID: 26027561 Review.
-
Adeno-associated virus (AAV) capsid engineering in liver-directed gene therapy.Expert Opin Biol Ther. 2021 Jun;21(6):749-766. doi: 10.1080/14712598.2021.1865303. Epub 2020 Dec 30. Expert Opin Biol Ther. 2021. PMID: 33331201 Review.
Cited by
-
Engineering Self-Assembling Protein Nanoparticles for Therapeutic Delivery.Bioconjug Chem. 2022 Nov 16;33(11):2018-2034. doi: 10.1021/acs.bioconjchem.2c00030. Epub 2022 Apr 29. Bioconjug Chem. 2022. PMID: 35487503 Free PMC article. Review.
-
Modeling reveals the strength of weak interactions in stacked-ring assembly.Biophys J. 2024 Jul 2;123(13):1763-1780. doi: 10.1016/j.bpj.2024.05.015. Epub 2024 May 18. Biophys J. 2024. PMID: 38762753 Free PMC article.
-
Gene Therapy in Hemophilia: Recent Advances.Int J Mol Sci. 2021 Jul 17;22(14):7647. doi: 10.3390/ijms22147647. Int J Mol Sci. 2021. PMID: 34299267 Free PMC article. Review.
-
Rational engineering of a functional CpG-free ITR for AAV gene therapy.Gene Ther. 2022 Jun;29(6):333-345. doi: 10.1038/s41434-021-00296-0. Epub 2021 Oct 6. Gene Ther. 2022. PMID: 34611321 Free PMC article.
-
Engineered compact pan-neuronal promoter from Alphaherpesvirus LAP2 enhances target gene expression in the mouse brain and reduces tropism in the liver.Gene Ther. 2024 May;31(5-6):335-344. doi: 10.1038/s41434-023-00430-0. Epub 2023 Nov 27. Gene Ther. 2024. PMID: 38012300 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
