Glutamate Signaling via the AMPAR Subunit GluR4 Regulates Oligodendrocyte Progenitor Cell Migration in the Developing Spinal Cord
- PMID: 33975920
- PMCID: PMC8221590
- DOI: 10.1523/JNEUROSCI.2562-20.2021
Glutamate Signaling via the AMPAR Subunit GluR4 Regulates Oligodendrocyte Progenitor Cell Migration in the Developing Spinal Cord
Abstract
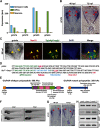
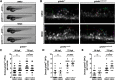
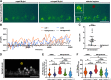
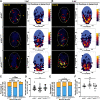
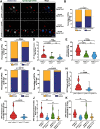
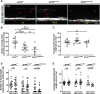
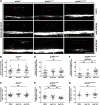
Oligodendrocyte progenitor cells (OPCs) are specified from discrete precursor populations during gliogenesis and migrate extensively from their origins, ultimately distributing throughout the brain and spinal cord during early development. Subsequently, a subset of OPCs differentiates into mature oligodendrocytes, which myelinate axons. This process is necessary for efficient neuronal signaling and organism survival. Previous studies have identified several factors that influence OPC development, including excitatory glutamatergic synapses that form between neurons and OPCs during myelination. However, little is known about how glutamate signaling affects OPC migration before myelination. In this study, we use in vivo, time-lapse imaging in zebrafish in conjunction with genetic and pharmacological perturbation to investigate OPC migration and myelination when the GluR4A ionotropic glutamate receptor subunit is disrupted. In our studies, we observed that gria4a mutant embryos and larvae displayed abnormal OPC migration and altered dorsoventral distribution in the spinal cord. Genetic mosaic analysis confirmed that these effects were cell-autonomous, and we identified that voltage-gated calcium channels were downstream of glutamate receptor signaling in OPCs and could rescue the migration and myelination defects we observed when glutamate signaling was perturbed. These results offer new insights into the complex system of neuron-OPC interactions and reveal a cell-autonomous role for glutamatergic signaling in OPCs during neural development.SIGNIFICANCE STATEMENT The migration of oligodendrocyte progenitor cells (OPCs) is an essential process during development that leads to uniform oligodendrocyte distribution and sufficient myelination for central nervous system function. Here, we demonstrate that the AMPA receptor (AMPAR) subunit GluR4A is an important driver of OPC migration and myelination in vivo and that activated voltage-gated calcium channels are downstream of glutamate receptor signaling in mediating this migration.
Keywords: glutamate signaling; myelin; oligodendrocyte; oligodendrocyte progenitor cell; zebrafish.
Copyright © 2021 the authors.
Figures



Similar articles
-
Myelin Proteolipid Protein Complexes with αv Integrin and AMPA Receptors In Vivo and Regulates AMPA-Dependent Oligodendrocyte Progenitor Cell Migration through the Modulation of Cell-Surface GluR2 Expression.J Neurosci. 2015 Aug 26;35(34):12018-32. doi: 10.1523/JNEUROSCI.5151-14.2015. J Neurosci. 2015. PMID: 26311781 Free PMC article.
-
In Vivo Regulation of Oligodendrocyte Precursor Cell Proliferation and Differentiation by the AMPA-Receptor Subunit GluA2.Cell Rep. 2018 Oct 23;25(4):852-861.e7. doi: 10.1016/j.celrep.2018.09.066. Cell Rep. 2018. PMID: 30355492
-
mTORC2 Loss in Oligodendrocyte Progenitor Cells Results in Regional Hypomyelination in the Central Nervous System.J Neurosci. 2023 Jan 25;43(4):540-558. doi: 10.1523/JNEUROSCI.0010-22.2022. Epub 2022 Dec 2. J Neurosci. 2023. PMID: 36460463 Free PMC article.
-
Mechanisms of oligodendrocyte progenitor developmental migration.Dev Neurobiol. 2021 Nov;81(8):985-996. doi: 10.1002/dneu.22856. Epub 2021 Oct 24. Dev Neurobiol. 2021. PMID: 34643996 Review.
-
Engineering biomaterial microenvironments to promote myelination in the central nervous system.Brain Res Bull. 2019 Oct;152:159-174. doi: 10.1016/j.brainresbull.2019.07.013. Epub 2019 Jul 12. Brain Res Bull. 2019. PMID: 31306690 Review.
Cited by
-
Glial Patchwork: Oligodendrocyte Progenitor Cells and Astrocytes Blanket the Central Nervous System.Front Cell Neurosci. 2022 Jan 5;15:803057. doi: 10.3389/fncel.2021.803057. eCollection 2021. Front Cell Neurosci. 2022. PMID: 35069117 Free PMC article. Review.
-
The neuropathobiology of multiple sclerosis.Nat Rev Neurosci. 2024 Jul;25(7):493-513. doi: 10.1038/s41583-024-00823-z. Epub 2024 May 24. Nat Rev Neurosci. 2024. PMID: 38789516 Review.
-
Roles of Ca2+ activity in injury-induced migration of microglia in zebrafish in vivo.Biochem Biophys Rep. 2022 Sep 9;32:101340. doi: 10.1016/j.bbrep.2022.101340. eCollection 2022 Dec. Biochem Biophys Rep. 2022. PMID: 36120493 Free PMC article.
-
Neuron to Oligodendrocyte Precursor Cell Synapses: Protagonists in Oligodendrocyte Development and Myelination, and Targets for Therapeutics.Front Neurosci. 2022 Jan 18;15:779125. doi: 10.3389/fnins.2021.779125. eCollection 2021. Front Neurosci. 2022. PMID: 35115904 Free PMC article. Review.
-
Recent Insights into the Functional Role of AMPA Receptors in the Oligodendrocyte Lineage Cells In Vivo.Int J Mol Sci. 2023 Feb 18;24(4):4138. doi: 10.3390/ijms24044138. Int J Mol Sci. 2023. PMID: 36835546 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
