Identification and characterisation of a phospholipid scramblase in the malaria parasite Plasmodium falciparum
- PMID: 33974939
- PMCID: PMC8202325
- DOI: 10.1016/j.molbiopara.2021.111374
Identification and characterisation of a phospholipid scramblase in the malaria parasite Plasmodium falciparum
Abstract
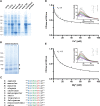
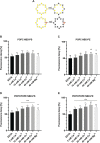
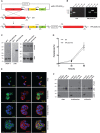
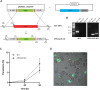
Recent studies highlight the emerging role of lipids as important messengers in malaria parasite biology. In an attempt to identify interacting proteins and regulators of these dynamic and versatile molecules, we hypothesised the involvement of phospholipid translocases and their substrates in the infection of the host erythrocyte by the malaria parasite Plasmodium spp. Here, using a data base searching approach of the Plasmodium Genomics Resources (www.plasmodb.org), we have identified a putative phospholipid (PL) scramblase in P. falciparum (PfPLSCR) that is conserved across the genus and in closely related unicellular algae. By reconstituting recombinant PfPLSCR into liposomes, we demonstrate metal ion dependent PL translocase activity and substrate preference, confirming PfPLSCR as a bona fide scramblase. We show that PfPLSCR is expressed during asexual and sexual parasite development, localising to different membranous compartments of the parasite throughout the intra-erythrocytic life cycle. Two different gene knockout approaches, however, suggest that PfPLSCR is not essential for erythrocyte invasion and asexual parasite development, pointing towards a possible role in other stages of the parasite life cycle.
Keywords: Gametocytes; Invasion; Liposomes; Malaria; Phospholipid scramblase; Plasmodium falciparum.
Copyright © 2021 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
Figures


Similar articles
-
Identification of a Plasmodium falciparum phospholipid transfer protein.J Biol Chem. 2013 Nov 1;288(44):31971-83. doi: 10.1074/jbc.M113.474189. Epub 2013 Sep 16. J Biol Chem. 2013. PMID: 24043620 Free PMC article.
-
Identification and characterization of DNA endonucleases in Plasmodium falciparum 3D7 clone.Malar J. 2018 Jun 18;17(1):232. doi: 10.1186/s12936-018-2388-0. Malar J. 2018. PMID: 29914511 Free PMC article.
-
Regulation and Essentiality of the StAR-related Lipid Transfer (START) Domain-containing Phospholipid Transfer Protein PFA0210c in Malaria Parasites.J Biol Chem. 2016 Nov 11;291(46):24280-24292. doi: 10.1074/jbc.M116.740506. Epub 2016 Oct 2. J Biol Chem. 2016. PMID: 27694132 Free PMC article.
-
Host cell invasion by malaria parasites.Parasitol Today. 2000 Oct;16(10):411-5. doi: 10.1016/s0169-4758(00)01756-7. Parasitol Today. 2000. PMID: 11006471 Review.
-
Molecules on the surface of the Plasmodium falciparum infected erythrocyte and their role in malaria pathogenesis and immune evasion.Mol Biochem Parasitol. 2001 Jul;115(2):129-43. doi: 10.1016/s0166-6851(01)00275-4. Mol Biochem Parasitol. 2001. PMID: 11420100 Review.
Cited by
-
Breakdown in membrane asymmetry regulation leads to monocyte recognition of P. falciparum-infected red blood cells.PLoS Pathog. 2021 Feb 18;17(2):e1009259. doi: 10.1371/journal.ppat.1009259. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33600495 Free PMC article.
-
Plasmodium sporozoite phospholipid scramblase interacts with mammalian carbamoyl-phosphate synthetase 1 to infect hepatocytes.Nat Commun. 2021 Nov 19;12(1):6773. doi: 10.1038/s41467-021-27109-7. Nat Commun. 2021. PMID: 34799567 Free PMC article.
-
Of membranes and malaria: phospholipid asymmetry in Plasmodium falciparum-infected red blood cells.Cell Mol Life Sci. 2021 May;78(10):4545-4561. doi: 10.1007/s00018-021-03799-6. Epub 2021 Mar 13. Cell Mol Life Sci. 2021. PMID: 33713154 Free PMC article. Review.
-
The enemy within: lipid asymmetry in intracellular parasite-host interactions.Emerg Top Life Sci. 2023 Mar 31;7(1):67-79. doi: 10.1042/ETLS20220089. Emerg Top Life Sci. 2023. PMID: 36820809 Free PMC article. Review.
-
Targeting Plasmodium Life Cycle with Novel Parasite Ligands as Vaccine Antigens.Vaccines (Basel). 2024 Apr 30;12(5):484. doi: 10.3390/vaccines12050484. Vaccines (Basel). 2024. PMID: 38793735 Free PMC article. Review.
References
-
- Vial H.J., Eldin P., Tielens A.G.M., van Hellemond J.J. Phospholipids in parasitic protozoa. Mol. Biochem. Parasitol. 2003;126:143–154. - PubMed
-
- Gulati S., Ekland E.H., Ruggles K.V., Chan R.B., Jayabalasingham B., Zhou B., Mantel P.Y., Lee M.C., Spottiswoode N., Coburn-Flynn O. Profiling the essential nature of lipid metabolism in asexual blood and gametocyte stages of plasmodium falciparum. Cell Host Microbe. 2015;18:371–381. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

