Sumoylation of the Carboxy-Terminal of Human Cytomegalovirus DNA Polymerase Processivity Factor UL44 Attenuates Viral DNA Replication
- PMID: 33967989
- PMCID: PMC8097051
- DOI: 10.3389/fmicb.2021.652719
Sumoylation of the Carboxy-Terminal of Human Cytomegalovirus DNA Polymerase Processivity Factor UL44 Attenuates Viral DNA Replication
Abstract
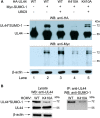
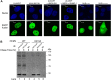
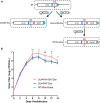
Controlled regulation of genomic DNA synthesis is a universally conserved process for all herpesviruses, including human cytomegalovirus (HCMV), and plays a key role in viral pathogenesis, such as persistent infections. HCMV DNA polymerase processivity factor UL44 plays an essential role in viral DNA replication. To better understand the biology of UL44, we performed a yeast two-hybrid screen for host proteins that could interact with UL44. The most frequently isolated result was the SUMO-conjugating enzyme UBC9, a protein involved in the sumoylation pathway. The UBC9-UL44 interaction was confirmed by in vitro His-tag pull-down and in vivo co-immunoprecipitation assays. Using deletion mutants of UL44, we mapped two small regions of UL44, aa 11-16, and 260-269, which might be critical for the interaction with UBC9. We then demonstrated that UL44 was a target for sumoylation by in vitro and in vivo sumoylation assays, as well as in HCMV-infected cells. We further confirmed that 410lysine located within a ψKxE consensus motif on UL44 carboxy-terminal was the major sumoylation site of UL44. Interestingly, although 410lysine had no effects on subcellular localization or protein stability of UL44, the removal of 410lysine sumoylation site enhanced both viral DNA synthesis in transfection-replication assays and viral progeny production in infected cells for HCMV, suggesting sumoylation can attenuate HCMV replication through targeting UL44. Our results suggest that sumoylation plays a key role in regulating UL44 functions and viral replication, and reveal the crucial role of the carboxy-terminal of UL44, for which little function has been known before.
Keywords: DNA replication; UBC9; UL44; human cytomegalovirus; sumoylation.
Copyright © 2021 Chen, Li, He, Li, Niu, Cao and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



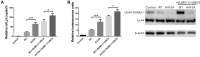

Similar articles
-
The SUMOylation of Human Cytomegalovirus Capsid Assembly Protein Precursor (UL80.5) Affects Its Interaction with Major Capsid Protein (UL86) and Viral Replication.Viruses. 2023 Apr 7;15(4):931. doi: 10.3390/v15040931. Viruses. 2023. PMID: 37112911 Free PMC article.
-
Site-specific SUMOylation of viral polymerase processivity factor: a way of localizingtoND10 subnuclear domains for restricted and self-controlled reproduction of herpesvirus.Virulence. 2021 Dec;12(1):2883-2901. doi: 10.1080/21505594.2021.2000689. Virulence. 2021. PMID: 34747321 Free PMC article.
-
The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner.PLoS One. 2012;7(11):e49630. doi: 10.1371/journal.pone.0049630. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166733 Free PMC article.
-
Role of the specific interaction of UL112-113 p84 with UL44 DNA polymerase processivity factor in promoting DNA replication of human cytomegalovirus.J Virol. 2010 Sep;84(17):8409-21. doi: 10.1128/JVI.00189-10. Epub 2010 Jun 10. J Virol. 2010. PMID: 20538862 Free PMC article.
-
SUMO Ubc9 enzyme as a viral target.IUBMB Life. 2014 Jan;66(1):27-33. doi: 10.1002/iub.1240. Epub 2014 Jan 6. IUBMB Life. 2014. PMID: 24395713 Review.
Cited by
-
SUMOylation and Viral Infections of the Brain.Pathogens. 2022 Jul 21;11(7):818. doi: 10.3390/pathogens11070818. Pathogens. 2022. PMID: 35890062 Free PMC article. Review.
-
A UL26-PIAS1 complex antagonizes anti-viral gene expression during Human Cytomegalovirus infection.PLoS Pathog. 2024 May 20;20(5):e1012058. doi: 10.1371/journal.ppat.1012058. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38768227 Free PMC article.
-
SUMOylation in Viral Replication and Antiviral Defense.Adv Sci (Weinh). 2022 Mar;9(7):e2104126. doi: 10.1002/advs.202104126. Epub 2022 Jan 21. Adv Sci (Weinh). 2022. PMID: 35060688 Free PMC article. Review.
-
The SUMOylation of Human Cytomegalovirus Capsid Assembly Protein Precursor (UL80.5) Affects Its Interaction with Major Capsid Protein (UL86) and Viral Replication.Viruses. 2023 Apr 7;15(4):931. doi: 10.3390/v15040931. Viruses. 2023. PMID: 37112911 Free PMC article.
-
Insights into the Transcriptome of Human Cytomegalovirus: A Comprehensive Review.Viruses. 2023 Aug 8;15(8):1703. doi: 10.3390/v15081703. Viruses. 2023. PMID: 37632045 Free PMC article. Review.
References
-
- Ahn J. H., Xu Y., Jang W. J., Matunis M. J., Hayward G. S. (2001). Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75 3859–3872. 10.1128/jvi.75.8.3859-3872.2001 - DOI - PMC - PubMed
-
- Appleton B. A., Brooks J., Loregian A., Filman D. J., Coen D. M., Hogle J. M. (2006). Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 inΔ3, with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44. J. Biol. Chem. 281 5224–5232. 10.1074/jbc.m506900200 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

