Foxo1-induced miR-92b down-regulation promotes blood-brain barrier damage after ischaemic stroke by targeting NOX4
- PMID: 33955666
- PMCID: PMC8178288
- DOI: 10.1111/jcmm.16537
Foxo1-induced miR-92b down-regulation promotes blood-brain barrier damage after ischaemic stroke by targeting NOX4
Abstract
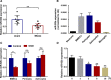
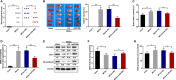
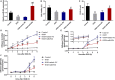
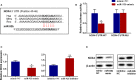
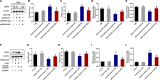
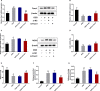
The blood-brain barrier (BBB) damage is a momentous pathological process of ischaemic stroke. NADPH oxidases 4 (NOX4) boosts BBB damage after ischaemic stroke and its expression can be influenced by microRNAs. This study aimed to probe into whether miR-92b influenced the BBB damage after ischaemic stroke by regulating NOX4 expression. Here, miR-92b expression was lessened in the ischaemic brains of rats and oxygen-glucose deprivation (OGD)-induced brain microvascular endothelial cells (BMECs). In middle cerebral artery occlusion (MCAo) rats, miR-92b overexpression relieved the ameliorated neurological function and protected the BBB integrity. In vitro model, miR-92b overexpression raised the viability and lessened the permeability of OGD-induced BMECs. miR-92b targeted NOX4 and regulated the viability and permeability of OGD-induced BMECs by negatively modulating NOX4 expression. The transcription factor Foxo1 bound to the miR-92b promoter and restrained its expression. Foxo1 expression was induced by OGD-induction and its knockdown abolished the effects of OGD on miR-92b and NOX4 expressions, cell viability and permeability of BMECs. In general, our findings expounded that Foxo1-induced lessening miR-92b boosted BBB damage after ischaemic stroke by raising NOX4 expression.
Keywords: Foxo1; NOX4; blood-brain barrier; ischaemic stroke; miR-92b.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
All authors declare that they have no conflicts of interest in this work.
Figures

Similar articles
-
LncRNA SNHG8 sponges miR-449c-5p and regulates the SIRT1/FoxO1 pathway to affect microglia activation and blood-brain barrier permeability in ischemic stroke.J Leukoc Biol. 2022 May;111(5):953-966. doi: 10.1002/JLB.1A0421-217RR. Epub 2021 Sep 29. J Leukoc Biol. 2022. PMID: 34585441
-
MiR-92b-3p regulates oxygen and glucose deprivation-reperfusion-mediated apoptosis and inflammation by targeting TRAF3 in PC12 cells.Exp Physiol. 2020 Oct;105(10):1792-1801. doi: 10.1113/EP088708. Epub 2020 Sep 4. Exp Physiol. 2020. PMID: 32818322
-
Exosomal miR-370-3p increases the permeability of blood-brain barrier in ischemia/reperfusion stroke of brain by targeting MPK1.Aging (Albany NY). 2023 Mar 8;15(6):1931-1943. doi: 10.18632/aging.204573. Epub 2023 Mar 8. Aging (Albany NY). 2023. PMID: 37000151 Free PMC article.
-
Oxidative Injury in Ischemic Stroke: A Focus on NADPH Oxidase 4.Oxid Med Cell Longev. 2022 Feb 3;2022:1148874. doi: 10.1155/2022/1148874. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35154560 Free PMC article. Review.
-
Role of microRNA-34a in blood-brain barrier permeability and mitochondrial function in ischemic stroke.Front Cell Neurosci. 2023 Oct 19;17:1278334. doi: 10.3389/fncel.2023.1278334. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37927446 Free PMC article. Review.
Cited by
-
Ischemia Reperfusion Injury Induced Blood Brain Barrier Dysfunction and the Involved Molecular Mechanism.Neurochem Res. 2023 Aug;48(8):2320-2334. doi: 10.1007/s11064-023-03923-x. Epub 2023 Apr 5. Neurochem Res. 2023. PMID: 37017889 Review.
-
Neuroprotective Effect of Benzyl Ferulate on Ischemia/Reperfusion Injury via Regulating NOX2 and NOX4 in Rats: A Potential Antioxidant for CI/R Injury.Adv Pharmacol Pharm Sci. 2024 Nov 2;2024:5534135. doi: 10.1155/2024/5534135. eCollection 2024. Adv Pharmacol Pharm Sci. 2024. PMID: 39524533 Free PMC article.
-
Transcriptomic Profile of Blood-Brain Barrier Remodeling in Cerebral Amyloid Angiopathy.Front Cell Neurosci. 2022 Jun 22;16:931247. doi: 10.3389/fncel.2022.931247. eCollection 2022. Front Cell Neurosci. 2022. PMID: 35813502 Free PMC article.
-
Pathogenesis and Therapeutic Targets of Focal Cortical Dysplasia Based on Bioinformatics Analysis.Neurochem Res. 2022 Nov;47(11):3506-3521. doi: 10.1007/s11064-022-03715-9. Epub 2022 Aug 9. Neurochem Res. 2022. PMID: 35945307
-
Role of NADPH Oxidases in Blood-Brain Barrier Disruption and Ischemic Stroke.Antioxidants (Basel). 2022 Sep 30;11(10):1966. doi: 10.3390/antiox11101966. Antioxidants (Basel). 2022. PMID: 36290688 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

