A parental transcriptional response to microsporidia infection induces inherited immunity in offspring
- PMID: 33952520
- PMCID: PMC8099193
- DOI: 10.1126/sciadv.abf3114
A parental transcriptional response to microsporidia infection induces inherited immunity in offspring
Abstract
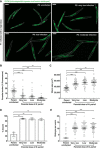
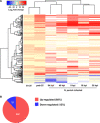
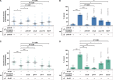
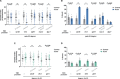
Parental infection can result in the production of offspring with enhanced immunity phenotypes. Critically, the mechanisms underlying inherited immunity are poorly understood. Here, we show that Caenorhabditis elegans infected with the intracellular microsporidian parasite N. parisii produce progeny that are resistant to microsporidia infection. We determine the kinetics of the response and show that intergenerational immunity prevents host-cell invasion by Nematocida parisii and enhances survival to the bacterial pathogen Pseudomonas aeruginosa We demonstrate that immunity is induced by the parental transcriptional response to infection, which can be mimicked through maternal somatic depletion of PALS-22 and the retinoblastoma protein ortholog, LIN-35. We find that other biotic and abiotic stresses (viral infection and cadmium exposure) that induce a similar transcriptional response as microsporidia also induce immunity in progeny. Together, our results reveal how a parental transcriptional signal can be induced by distinct stimuli and protect offspring against multiple classes of pathogens.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures




Similar articles
-
Studying Inherited Immunity in a Caenorhabditis elegans Model of Microsporidia Infection.J Vis Exp. 2022 Apr 6;(182). doi: 10.3791/63636. J Vis Exp. 2022. PMID: 35467660
-
PALS-14 promotes resistance to Nematocida parisii infection in Caenorhabditis elegans.MicroPubl Biol. 2024 Oct 15;2024:10.17912/micropub.biology.001325. doi: 10.17912/micropub.biology.001325. eCollection 2024. MicroPubl Biol. 2024. PMID: 39473452 Free PMC article.
-
Ubiquitin-mediated response to microsporidia and virus infection in C. elegans.PLoS Pathog. 2014 Jun 19;10(6):e1004200. doi: 10.1371/journal.ppat.1004200. eCollection 2014 Jun. PLoS Pathog. 2014. PMID: 24945527 Free PMC article.
-
Host-Microsporidia Interactions in Caenorhabditis elegans, a Model Nematode Host.Microbiol Spectr. 2016 Oct;4(5). doi: 10.1128/microbiolspec.FUNK-0003-2016. Microbiol Spectr. 2016. PMID: 27763260 Review.
-
Insights from C. elegans into Microsporidia Biology and Host-Pathogen Relationships.Exp Suppl. 2022;114:115-136. doi: 10.1007/978-3-030-93306-7_5. Exp Suppl. 2022. PMID: 35544001 Free PMC article. Review.
Cited by
-
Conservation of Nematocida microsporidia gene expression and host response in Caenorhabditis nematodes.PLoS One. 2022 Dec 19;17(12):e0279103. doi: 10.1371/journal.pone.0279103. eCollection 2022. PLoS One. 2022. PMID: 36534656 Free PMC article.
-
Genomic and phenotypic evolution of nematode-infecting microsporidia.PLoS Pathog. 2023 Jul 20;19(7):e1011510. doi: 10.1371/journal.ppat.1011510. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37471459 Free PMC article.
-
High-throughput small molecule screen identifies inhibitors of microsporidia invasion and proliferation in C. elegans.Nat Commun. 2022 Sep 26;13(1):5653. doi: 10.1038/s41467-022-33400-y. Nat Commun. 2022. PMID: 36163337 Free PMC article.
-
An organismal understanding of C. elegans innate immune responses, from pathogen recognition to multigenerational resistance.Semin Cell Dev Biol. 2024 Feb 15;154(Pt A):77-84. doi: 10.1016/j.semcdb.2023.03.005. Epub 2023 Mar 23. Semin Cell Dev Biol. 2024. PMID: 36966075 Free PMC article. Review.
-
Bacterial filamentation as a mechanism for cell-to-cell spread within an animal host.Nat Commun. 2022 Feb 4;13(1):693. doi: 10.1038/s41467-022-28297-6. Nat Commun. 2022. PMID: 35121734 Free PMC article.
References
-
- Little T. J., O’Connor B., Colegrave N., Watt K., Read A. F., Maternal transfer of strain-specific immunity in an invertebrate. Curr. Biol. 13, 489–492 (2003). - PubMed
-
- Norouzitallab P., Baruah K., Vandegehuchte M., Van Stappen G., Catania F., Bussche J. V., Vanhaecke L., Sorgeloos P., Bossier P., Environmental heat stress induces epigenetic transgenerational inheritance of robustness in parthenogenetic Artemia model. FASEB J. 28, 3552–3563 (2014). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

