The Dynamic SecYEG Translocon
- PMID: 33937339
- PMCID: PMC8082313
- DOI: 10.3389/fmolb.2021.664241
The Dynamic SecYEG Translocon
Abstract
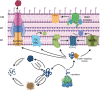
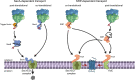
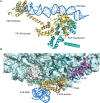
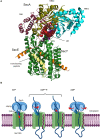
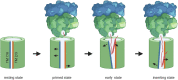
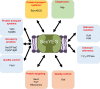
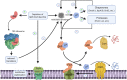
The spatial and temporal coordination of protein transport is an essential cornerstone of the bacterial adaptation to different environmental conditions. By adjusting the protein composition of extra-cytosolic compartments, like the inner and outer membranes or the periplasmic space, protein transport mechanisms help shaping protein homeostasis in response to various metabolic cues. The universally conserved SecYEG translocon acts at the center of bacterial protein transport and mediates the translocation of newly synthesized proteins into and across the cytoplasmic membrane. The ability of the SecYEG translocon to transport an enormous variety of different substrates is in part determined by its ability to interact with multiple targeting factors, chaperones and accessory proteins. These interactions are crucial for the assisted passage of newly synthesized proteins from the cytosol into the different bacterial compartments. In this review, we summarize the current knowledge about SecYEG-mediated protein transport, primarily in the model organism Escherichia coli, and describe the dynamic interaction of the SecYEG translocon with its multiple partner proteins. We furthermore highlight how protein transport is regulated and explore recent developments in using the SecYEG translocon as an antimicrobial target.
Keywords: FtsY; PpiD; SecA; SecYEG translocon; YidC; protein transport; signal recognition particle; stress response.
Copyright © 2021 Oswald, Njenga, Natriashvili, Sarmah and Koch.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

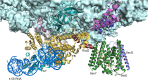


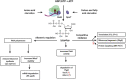
Similar articles
-
Noncompetitive binding of PpiD and YidC to the SecYEG translocon expands the global view on the SecYEG interactome in Escherichia coli.J Biol Chem. 2019 Dec 13;294(50):19167-19183. doi: 10.1074/jbc.RA119.010686. Epub 2019 Nov 7. J Biol Chem. 2019. PMID: 31699901 Free PMC article.
-
Co-translational protein targeting in bacteria.FEMS Microbiol Lett. 2018 Jun 1;365(11). doi: 10.1093/femsle/fny095. FEMS Microbiol Lett. 2018. PMID: 29790984 Review.
-
The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon.Biochemistry. 2008 May 20;47(20):5649-56. doi: 10.1021/bi800233w. Epub 2008 Apr 26. Biochemistry. 2008. PMID: 18439025
-
Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins.Mol Biol Cell. 2009 Mar;20(6):1804-15. doi: 10.1091/mbc.e08-08-0886. Epub 2009 Jan 21. Mol Biol Cell. 2009. PMID: 19158385 Free PMC article.
-
The Sec translocon mediated protein transport in prokaryotes and eukaryotes.Mol Membr Biol. 2014 Mar-May;31(2-3):58-84. doi: 10.3109/09687688.2014.907455. Mol Membr Biol. 2014. PMID: 24762201 Review.
Cited by
-
mRNA targeting eliminates the need for the signal recognition particle during membrane protein insertion in bacteria.Cell Rep. 2023 Mar 28;42(3):112140. doi: 10.1016/j.celrep.2023.112140. Epub 2023 Feb 25. Cell Rep. 2023. PMID: 36842086 Free PMC article.
-
The insertase YidC chaperones the polytopic membrane protein MelB inserting and folding simultaneously from both termini.Structure. 2023 Nov 2;31(11):1419-1430.e5. doi: 10.1016/j.str.2023.08.012. Epub 2023 Sep 13. Structure. 2023. PMID: 37708891 Free PMC article.
-
Methods to study folding of alpha-helical membrane proteins in lipids.Open Biol. 2022 Jul;12(7):220054. doi: 10.1098/rsob.220054. Epub 2022 Jul 20. Open Biol. 2022. PMID: 35855589 Free PMC article. Review.
-
Not sorcery after all: Roles of multiple charged residues in membrane insertion of gasdermin-A3.Front Cell Dev Biol. 2022 Sep 2;10:958957. doi: 10.3389/fcell.2022.958957. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36120563 Free PMC article.
-
Targeting the 'garbage-bin' to fight cancer: HDAC6 inhibitor WT161 has an anti-tumor effect on osteosarcoma and synergistically interacts with 5-FU.Biosci Rep. 2021 Aug 27;41(8):BSR20210952. doi: 10.1042/BSR20210952. Biosci Rep. 2021. PMID: 34323266 Free PMC article.
References
-
- Akiyama Y., Kihara A., Tokuda H., Ito K. (1996). FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 271 31196–31201. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

