Downregulating carnitine palmitoyl transferase 1 affects disease progression in the SOD1 G93A mouse model of ALS
- PMID: 33931719
- PMCID: PMC8087699
- DOI: 10.1038/s42003-021-02034-z
Downregulating carnitine palmitoyl transferase 1 affects disease progression in the SOD1 G93A mouse model of ALS
Abstract
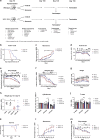
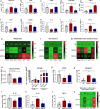
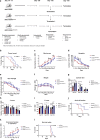
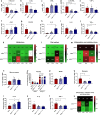
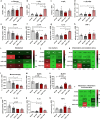
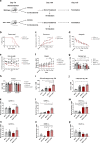
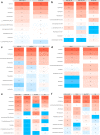
Amyotrophic lateral sclerosis (ALS) is a fatal motor neuron disease characterized by death of motor neurons. The etiology and pathogenesis remains elusive despite decades of intensive research. Herein, we report that dysregulated metabolism plays a central role in the SOD1 G93A mouse model mimicking ALS. Specifically, we report that the activity of carnitine palmitoyl transferase 1 (CPT1) lipid metabolism is associated with disease progression. Downregulation of CPT1 activity by pharmacological and genetic methods results in amelioration of disease symptoms, inflammation, oxidative stress and mitochondrial function, whereas upregulation by high-fat diet or corticosterone results in a more aggressive disease progression. Finally, we show that downregulating CPT1 shifts the gut microbiota communities towards a protective phenotype in SOD1 G93A mice. These findings reveal that metabolism, and specifically CPT1 lipid metabolism plays a central role in the SOD1 G93A mouse model and shows that CPT1 might be a therapeutic target in ALS.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Dysregulation of metabolic pathways by carnitine palmitoyl-transferase 1 plays a key role in central nervous system disorders: experimental evidence based on animal models.Sci Rep. 2020 Sep 24;10(1):15583. doi: 10.1038/s41598-020-72638-8. Sci Rep. 2020. PMID: 32973137 Free PMC article.
-
In-vivo genetic ablation of metabotropic glutamate receptor type 5 slows down disease progression in the SOD1G93A mouse model of amyotrophic lateral sclerosis.Neurobiol Dis. 2019 Sep;129:79-92. doi: 10.1016/j.nbd.2019.05.007. Epub 2019 May 15. Neurobiol Dis. 2019. PMID: 31102766
-
Knocking down metabotropic glutamate receptor 1 improves survival and disease progression in the SOD1(G93A) mouse model of amyotrophic lateral sclerosis.Neurobiol Dis. 2014 Apr;64:48-59. doi: 10.1016/j.nbd.2013.11.006. Epub 2013 Dec 19. Neurobiol Dis. 2014. PMID: 24361555
-
PACAP signaling exerts opposing effects on neuroprotection and neuroinflammation during disease progression in the SOD1(G93A) mouse model of amyotrophic lateral sclerosis.Neurobiol Dis. 2013 Jun;54:32-42. doi: 10.1016/j.nbd.2013.02.010. Epub 2013 Mar 4. Neurobiol Dis. 2013. PMID: 23466699 Free PMC article.
-
A Gut Feeling in Amyotrophic Lateral Sclerosis: Microbiome of Mice and Men.Front Cell Infect Microbiol. 2022 Mar 11;12:839526. doi: 10.3389/fcimb.2022.839526. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35360111 Free PMC article. Review.
Cited by
-
Intrathecal Injection of the Secretome from ALS Motor Neurons Regulated for miR-124 Expression Prevents Disease Outcomes in SOD1-G93A Mice.Biomedicines. 2022 Aug 29;10(9):2120. doi: 10.3390/biomedicines10092120. Biomedicines. 2022. PMID: 36140218 Free PMC article.
-
Oxidized low-density lipoprotein changes the inflammatory status and metabolomics profiles in human and mouse macrophages and microglia.Heliyon. 2024 Mar 28;10(7):e28806. doi: 10.1016/j.heliyon.2024.e28806. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38617955 Free PMC article.
-
Rethinking neurodegenerative diseases: neurometabolic concept linking lipid oxidation to diseases in the central nervous system.Neural Regen Res. 2024 Jul 1;19(7):1437-1445. doi: 10.4103/1673-5374.387965. Epub 2023 Nov 8. Neural Regen Res. 2024. PMID: 38051885 Free PMC article.
-
Regulation of gut microbiome by ketogenic diet in neurodegenerative diseases: A molecular crosstalk.Front Aging Neurosci. 2022 Oct 14;14:1015837. doi: 10.3389/fnagi.2022.1015837. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36313018 Free PMC article.
-
Repurposing of Trimetazidine for amyotrophic lateral sclerosis: A study in SOD1G93A mice.Br J Pharmacol. 2022 Apr;179(8):1732-1752. doi: 10.1111/bph.15738. Epub 2022 Jan 13. Br J Pharmacol. 2022. PMID: 34783031 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

