Binding Free Energy (BFE) Calculations and Quantitative Structure-Activity Relationship (QSAR) Analysis of Schistosoma mansoni Histone Deacetylase 8 (sm HDAC8) Inhibitors
- PMID: 33925246
- PMCID: PMC8125515
- DOI: 10.3390/molecules26092584
Binding Free Energy (BFE) Calculations and Quantitative Structure-Activity Relationship (QSAR) Analysis of Schistosoma mansoni Histone Deacetylase 8 (sm HDAC8) Inhibitors
Abstract
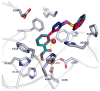
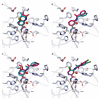
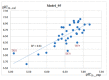
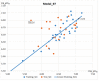
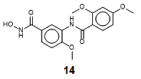
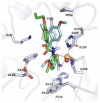
Histone-modifying proteins have been identified as promising targets to treat several diseases including cancer and parasitic ailments. In silico methods have been incorporated within a variety of drug discovery programs to facilitate the identification and development of novel lead compounds. In this study, we explore the binding modes of a series of benzhydroxamates derivatives developed as histone deacetylase inhibitors of Schistosoma mansoni histone deacetylase (smHDAC) using molecular docking and binding free energy (BFE) calculations. The developed docking protocol was able to correctly reproduce the experimentally established binding modes of resolved smHDAC8-inhibitor complexes. However, as has been reported in former studies, the obtained docking scores weakly correlate with the experimentally determined activity of the studied inhibitors. Thus, the obtained docking poses were refined and rescored using the Amber software. From the computed protein-inhibitor BFE, different quantitative structure-activity relationship (QSAR) models could be developed and validated using several cross-validation techniques. Some of the generated QSAR models with good correlation could explain up to ~73% variance in activity within the studied training set molecules. The best performing models were subsequently tested on an external test set of newly designed and synthesized analogs. In vitro testing showed a good correlation between the predicted and experimentally observed IC50 values. Thus, the generated models can be considered as interesting tools for the identification of novel smHDAC8 inhibitors.
Keywords: Schistosoma mansoni; binding free energy calculations; histone deacetylase inhibitors; quantitative structure–activity relationship (QSAR).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
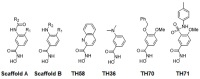
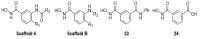

Similar articles
-
A Novel Class of Schistosoma mansoni Histone Deacetylase 8 (HDAC8) Inhibitors Identified by Structure-Based Virtual Screening and In Vitro Testing.Molecules. 2018 Mar 2;23(3):566. doi: 10.3390/molecules23030566. Molecules. 2018. PMID: 29498707 Free PMC article.
-
Discovery of inhibitors of Schistosoma mansoni HDAC8 by combining homology modeling, virtual screening, and in vitro validation.J Chem Inf Model. 2014 Oct 27;54(10):3005-19. doi: 10.1021/ci5004653. Epub 2014 Oct 2. J Chem Inf Model. 2014. PMID: 25243797
-
Molecular basis for the antiparasitic activity of a mercaptoacetamide derivative that inhibits histone deacetylase 8 (HDAC8) from the human pathogen schistosoma mansoni.J Mol Biol. 2014 Oct 9;426(20):3442-53. doi: 10.1016/j.jmb.2014.03.007. Epub 2014 Mar 20. J Mol Biol. 2014. PMID: 24657767
-
Drugging the schistosome zinc-dependent HDACs: current progress and future perspectives.Future Med Chem. 2015;7(6):783-800. doi: 10.4155/fmc.15.25. Future Med Chem. 2015. PMID: 25996070 Review.
-
Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases.Curr Opin Struct Biol. 2019 Dec;59:9-18. doi: 10.1016/j.sbi.2019.01.004. Epub 2019 Feb 8. Curr Opin Struct Biol. 2019. PMID: 30743180 Free PMC article. Review.
Cited by
-
Docking, Binding Free Energy Calculations and In Vitro Characterization of Pyrazine Linked 2-Aminobenzamides as Novel Class I Histone Deacetylase (HDAC) Inhibitors.Molecules. 2022 Apr 14;27(8):2526. doi: 10.3390/molecules27082526. Molecules. 2022. PMID: 35458724 Free PMC article.
-
Design, synthesis, and biochemical and computational screening of novel oxindole derivatives as inhibitors of Aurora A kinase and SARS-CoV-2 spike/host ACE2 interaction.Med Chem Res. 2024;33(4):620-634. doi: 10.1007/s00044-024-03201-7. Epub 2024 Mar 5. Med Chem Res. 2024. PMID: 38646411 Free PMC article.
-
Computational discovery of dual potential inhibitors of SARS-CoV-2 spike/ACE2 and Mpro: 3D-pharmacophore, docking-based virtual screening, quantum mechanics and molecular dynamics.Eur Biophys J. 2024 Aug;53(5-6):277-298. doi: 10.1007/s00249-024-01713-z. Epub 2024 Jun 21. Eur Biophys J. 2024. PMID: 38907013
-
Effects of some anti-ulcer and anti-inflammatory natural products on cyclooxygenase and lipoxygenase enzymes: insights from in silico analysis.In Silico Pharmacol. 2024 Nov 2;12(2):97. doi: 10.1007/s40203-024-00269-2. eCollection 2024. In Silico Pharmacol. 2024. PMID: 39498163
-
Histone Deacetylase (HDAC) Inhibitors for the Treatment of Schistosomiasis.Pharmaceuticals (Basel). 2022 Jan 10;15(1):80. doi: 10.3390/ph15010080. Pharmaceuticals (Basel). 2022. PMID: 35056137 Free PMC article. Review.
References
-
- World Health Organization Schistosomiasis Facts Sheet. [(accessed on 18 November 2020)];2018 Available online: https://www.who.int/news-room/fact-sheets/detail/schistosomiasis.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

