Identification of Novel Candidate Genes and Variants for Hearing Loss and Temporal Bone Anomalies
- PMID: 33924653
- PMCID: PMC8069784
- DOI: 10.3390/genes12040566
Identification of Novel Candidate Genes and Variants for Hearing Loss and Temporal Bone Anomalies
Abstract
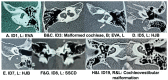
Background: Hearing loss remains an important global health problem that is potentially addressed through early identification of a genetic etiology, which helps to predict outcomes of hearing rehabilitation such as cochlear implantation and also to mitigate the long-term effects of comorbidities. The identification of variants for hearing loss and detailed descriptions of clinical phenotypes in patients from various populations are needed to improve the utility of clinical genetic screening for hearing loss. Methods: Clinical and exome data from 15 children with hearing loss were reviewed. Standard tools for annotating variants were used and rare, putatively deleterious variants were selected from the exome data. Results: In 15 children, 21 rare damaging variants in 17 genes were identified, including: 14 known hearing loss or neurodevelopmental genes, 11 of which had novel variants; and three candidate genes IST1, CBLN3 and GDPD5, two of which were identified in children with both hearing loss and enlarged vestibular aqueducts. Patients with variants within IST1 and MYO18B had poorer outcomes after cochlear implantation. Conclusion: Our findings highlight the importance of identifying novel variants and genes in ethnic groups that are understudied for hearing loss.
Keywords: CBLN3; GDPD5; IST1; anomalies; cochlear implant; enlarged vestibular aqueduct; genetic testing; hearing loss; inner ear; malformations; temporal bone.
Conflict of interest statement
C.T. is an employee of MED-EL, but MED-EL had no role in the study design, data analysis and manuscript preparation. All authors declare no conflict of interest.
Figures
Similar articles
-
High prevalence of inner-ear and/or internal auditory canal malformations in children with unilateral sensorineural hearing loss.Int J Pediatr Otorhinolaryngol. 2013 Feb;77(2):228-32. doi: 10.1016/j.ijporl.2012.11.001. Epub 2012 Nov 30. Int J Pediatr Otorhinolaryngol. 2013. PMID: 23200870
-
Novel Variants in Hearing Loss Genes and Associations With Audiometric Thresholds in a Multi-ethnic Cohort of US Patients With Cochlear Implants.Otol Neurotol. 2020 Aug;41(7):978-985. doi: 10.1097/MAO.0000000000002671. Otol Neurotol. 2020. PMID: 32658404
-
The SLC26A4 c.706C>G (p.Leu236Val) Variant is a Frequent Cause of Hearing Impairment in Filipino Cochlear Implantees.Otol Neurotol. 2018 Sep;39(8):e726-e730. doi: 10.1097/MAO.0000000000001893. Otol Neurotol. 2018. PMID: 30113565 Free PMC article.
-
Aspects of temporal bone anatomy and pathology in conjunction with cochlear implant surgery.Acta Radiol Suppl. 2003 Jul;430:2-15. Acta Radiol Suppl. 2003. PMID: 12834396 Review.
-
The Importance of Early Genetic Diagnostics of Hearing Loss in Children.Medicina (Kaunas). 2020 Sep 14;56(9):471. doi: 10.3390/medicina56090471. Medicina (Kaunas). 2020. PMID: 32937936 Free PMC article. Review.
Cited by
-
Variants of human CLDN9 cause mild to profound hearing loss.Hum Mutat. 2021 Oct;42(10):1321-1335. doi: 10.1002/humu.24260. Epub 2021 Aug 1. Hum Mutat. 2021. PMID: 34265170 Free PMC article.
-
Newborn Hearing Screening and Beyond: A Continuing Journey in the Philippines.Acta Med Philipp. 2023 Sep 27;57(9):7-14. doi: 10.47895/amp.v57i9.8836. eCollection 2023. Acta Med Philipp. 2023. PMID: 39483801 Free PMC article. No abstract available.
-
The genome of the black-footed cat: Revealing a rich natural history and urgent conservation priorities for small felids.Proc Natl Acad Sci U S A. 2024 Jan 9;121(2):e2310763120. doi: 10.1073/pnas.2310763120. Epub 2024 Jan 2. Proc Natl Acad Sci U S A. 2024. PMID: 38165928 Free PMC article.
-
The role of SLC12A family of cation-chloride cotransporters and drug discovery methodologies.J Pharm Anal. 2023 Dec;13(12):1471-1495. doi: 10.1016/j.jpha.2023.09.002. Epub 2023 Sep 9. J Pharm Anal. 2023. PMID: 38223443 Free PMC article. Review.
-
Advanced Omics Techniques for Understanding Cochlear Genome, Epigenome, and Transcriptome in Health and Disease.Biomolecules. 2023 Oct 17;13(10):1534. doi: 10.3390/biom13101534. Biomolecules. 2023. PMID: 37892216 Free PMC article. Review.
References
-
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. - DOI - PMC - PubMed
-
- Azaiez H., Booth K.T., Ephraim S.S., Crone B., Black-Ziegelbein E.A., Marini R.J., Shearer A.E., Sloan-Heggen C.M., Kolbe D., Casavant T., et al. Genomic landscape and mutational signatures of deafness-associated genes. Am. J. Hum. Genet. 2018;103:484–497. doi: 10.1016/j.ajhg.2018.08.006. - DOI - PMC - PubMed
-
- Girotto G., Morgan A., Krishnamoorthy N., Cocca M., Brumat M., Bassani S., La Bianca M., Di Stazio M., Gasparini P. Next generation sequencing and animal models reveal SLC9A3R1 as a new gene involved in human age-related hearing loss. Front. Genet. 2020;10:142. doi: 10.3389/fgene.2019.00142. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

