RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases
- PMID: 33922187
- PMCID: PMC8145807
- DOI: 10.3390/genes12050627
RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases
Abstract
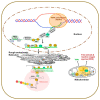
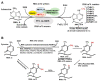
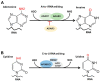
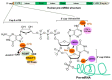
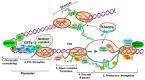
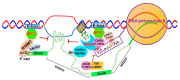
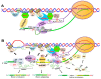
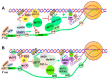
Chromatin structure plays an essential role in eukaryotic gene expression and cell identity. Traditionally, DNA and histone modifications have been the focus of chromatin regulation; however, recent molecular and imaging studies have revealed an intimate connection between RNA epigenetics and chromatin structure. Accumulating evidence suggests that RNA serves as the interplay between chromatin and the transcription and splicing machineries within the cell. Additionally, epigenetic modifications of nascent RNAs fine-tune these interactions to regulate gene expression at the co- and post-transcriptional levels in normal cell development and human diseases. This review will provide an overview of recent advances in the emerging field of RNA epigenetics, specifically the role of RNA modifications and RNA modifying proteins in chromatin remodeling, transcription activation and RNA processing, as well as translational implications in human diseases.
Keywords: 2’-O-methylation (Nm); 5-methylcytosine (m5C); 5’ cap (5’ cap); 7-methylguanosine (m7G); A-to-I; C-to-U; MYC; N6-methyladenosine (m6A); NOL1/NOP2/sun domain (NSUN); R-loops; RNA editing.
Conflict of interest statement
No conflicts of interest declared by the authors.
Figures
Similar articles
-
The Role of RNA Modifications and RNA-modifying Proteins in Cancer Therapy and Drug Resistance.Curr Cancer Drug Targets. 2021;21(4):326-352. doi: 10.2174/1568009621666210127092828. Curr Cancer Drug Targets. 2021. PMID: 33504307 Review.
-
RNA Modifications and Epigenetics in Modulation of Lung Cancer and Pulmonary Diseases.Int J Mol Sci. 2021 Sep 30;22(19):10592. doi: 10.3390/ijms221910592. Int J Mol Sci. 2021. PMID: 34638933 Free PMC article. Review.
-
The crosstalk between m6A RNA methylation and other epigenetic regulators: a novel perspective in epigenetic remodeling.Theranostics. 2021 Mar 4;11(9):4549-4566. doi: 10.7150/thno.54967. eCollection 2021. Theranostics. 2021. PMID: 33754077 Free PMC article. Review.
-
Versatile functions of RNA m6A machinery on chromatin.J Mol Cell Biol. 2022 Jul 8;14(3):mjac011. doi: 10.1093/jmcb/mjac011. J Mol Cell Biol. 2022. PMID: 35212732 Free PMC article.
-
Investigation of epigenetics in kidney cell biology.Methods Cell Biol. 2019;153:255-278. doi: 10.1016/bs.mcb.2019.04.015. Epub 2019 Jun 13. Methods Cell Biol. 2019. PMID: 31395383 Free PMC article. Review.
Cited by
-
Whole Exome Sequencing in a Population With Severe Congenital Anomalies of Kidney and Urinary Tract.Front Pediatr. 2022 Aug 4;10:898773. doi: 10.3389/fped.2022.898773. eCollection 2022. Front Pediatr. 2022. PMID: 35990004 Free PMC article.
-
Exploring the Epigenetic Regulatory Role of m6A-Associated SNPs in Type 2 Diabetes Pathogenesis.Pharmgenomics Pers Med. 2021 Oct 27;14:1369-1378. doi: 10.2147/PGPM.S334346. eCollection 2021. Pharmgenomics Pers Med. 2021. PMID: 34737607 Free PMC article.
-
Genome-wide detection of m6A-associated SNPs in atrial fibrillation pathogenesis.Front Cardiovasc Med. 2023 May 26;10:1152851. doi: 10.3389/fcvm.2023.1152851. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 37304952 Free PMC article.
-
The Methylation Game: Epigenetic and Epitranscriptomic Dynamics of 5-Methylcytosine.Front Cell Dev Biol. 2022 Jun 3;10:915685. doi: 10.3389/fcell.2022.915685. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35721489 Free PMC article. Review.
-
The nuclear and cytoplasmic activities of RNA polymerase III, and an evolving transcriptome for surveillance.Nucleic Acids Res. 2021 Dec 2;49(21):12017-12034. doi: 10.1093/nar/gkab1145. Nucleic Acids Res. 2021. PMID: 34850129 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

