Retrotransposons as Drivers of Mammalian Brain Evolution
- PMID: 33922141
- PMCID: PMC8143547
- DOI: 10.3390/life11050376
Retrotransposons as Drivers of Mammalian Brain Evolution
Abstract
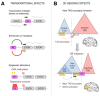
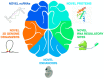
Retrotransposons, a large and diverse class of transposable elements that are still active in humans, represent a remarkable force of genomic innovation underlying mammalian evolution. Among the features distinguishing mammals from all other vertebrates, the presence of a neocortex with a peculiar neuronal organization, composition and connectivity is perhaps the one that, by affecting the cognitive abilities of mammals, contributed mostly to their evolutionary success. Among mammals, hominids and especially humans display an extraordinarily expanded cortical volume, an enrichment of the repertoire of neural cell types and more elaborate patterns of neuronal connectivity. Retrotransposon-derived sequences have recently been implicated in multiple layers of gene regulation in the brain, from transcriptional and post-transcriptional control to both local and large-scale three-dimensional chromatin organization. Accordingly, an increasing variety of neurodevelopmental and neurodegenerative conditions are being recognized to be associated with retrotransposon dysregulation. We review here a large body of recent studies lending support to the idea that retrotransposon-dependent evolutionary novelties were crucial for the emergence of mammalian, primate and human peculiarities of brain morphology and function.
Keywords: ERV; LINE-1; SINE; enhancer; exaptation; neocortex; neuron; noncoding RNA; transposable elements.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Similar Evolutionary Trajectories for Retrotransposon Accumulation in Mammals.Genome Biol Evol. 2017 Sep 1;9(9):2336-2353. doi: 10.1093/gbe/evx179. Genome Biol Evol. 2017. PMID: 28945883 Free PMC article.
-
DNA Editing of LTR Retrotransposons Reveals the Impact of APOBECs on Vertebrate Genomes.Mol Biol Evol. 2016 Feb;33(2):554-67. doi: 10.1093/molbev/msv239. Epub 2015 Nov 4. Mol Biol Evol. 2016. PMID: 26541172 Free PMC article.
-
Mammalian transposable elements and their impacts on genome evolution.Chromosome Res. 2018 Mar;26(1-2):25-43. doi: 10.1007/s10577-017-9570-z. Epub 2018 Feb 1. Chromosome Res. 2018. PMID: 29392473 Free PMC article. Review.
-
Control of mammalian retrotransposons by cellular RNA processing activities.Mob Genet Elements. 2014 Mar 6;4:e28439. doi: 10.4161/mge.28439. eCollection 2014. Mob Genet Elements. 2014. PMID: 25346866 Free PMC article.
-
Genetic Mechanisms Underlying Cortical Evolution in Mammals.Front Cell Dev Biol. 2021 Feb 15;9:591017. doi: 10.3389/fcell.2021.591017. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33659245 Free PMC article. Review.
Cited by
-
HERV-K(HML7) Integrations in the Human Genome: Comprehensive Characterization and Comparative Analysis in Non-Human Primates.Biology (Basel). 2021 May 14;10(5):439. doi: 10.3390/biology10050439. Biology (Basel). 2021. PMID: 34069102 Free PMC article.
-
Evolutionarily recent retrotransposons contribute to schizophrenia.Res Sq [Preprint]. 2023 Jan 23:rs.3.rs-2474682. doi: 10.21203/rs.3.rs-2474682/v1. Res Sq. 2023. Update in: Transl Psychiatry. 2023 May 27;13(1):181. doi: 10.1038/s41398-023-02472-9 PMID: 36747630 Free PMC article. Updated. Preprint.
-
Human Endogenous Retrovirus (HERV) Transcriptome Is Dynamically Modulated during SARS-CoV-2 Infection and Allows Discrimination of COVID-19 Clinical Stages.Microbiol Spectr. 2023 Feb 14;11(1):e0251622. doi: 10.1128/spectrum.02516-22. Epub 2023 Jan 5. Microbiol Spectr. 2023. PMID: 36602345 Free PMC article.
-
The Role of Transposable Elements of the Human Genome in Neuronal Function and Pathology.Int J Mol Sci. 2022 May 23;23(10):5847. doi: 10.3390/ijms23105847. Int J Mol Sci. 2022. PMID: 35628657 Free PMC article. Review.
-
Transcriptomics of Parental Care in the Hypothalamic-Septal Region of Female Zebra Finch Brain.Int J Mol Sci. 2022 Feb 24;23(5):2518. doi: 10.3390/ijms23052518. Int J Mol Sci. 2022. PMID: 35269661 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

