Functional Interfaces, Biological Pathways, and Regulations of Interferon-Related DNA Damage Resistance Signature (IRDS) Genes
- PMID: 33922087
- PMCID: PMC8143464
- DOI: 10.3390/biom11050622
Functional Interfaces, Biological Pathways, and Regulations of Interferon-Related DNA Damage Resistance Signature (IRDS) Genes
Abstract
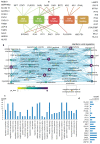
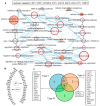
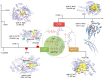
Interferon (IFN)-related DNA damage resistant signature (IRDS) genes are a subgroup of interferon-stimulated genes (ISGs) found upregulated in different cancer types, which promotes resistance to DNA damaging chemotherapy and radiotherapy. Along with briefly discussing IFNs and signalling in this review, we highlighted how different IRDS genes are affected by viruses. On the contrary, different strategies adopted to suppress a set of IRDS genes (STAT1, IRF7, OAS family, and BST2) to induce (chemo- and radiotherapy) sensitivity were deliberated. Significant biological pathways that comprise these genes were classified, along with their frequently associated genes (IFIT1/3, IFITM1, IRF7, ISG15, MX1/2 and OAS1/3/L). Major upstream regulators from the IRDS genes were identified, and different IFN types regulating these genes were outlined. Functional interfaces of IRDS proteins with DNA/RNA/ATP/GTP/NADP biomolecules featured a well-defined pharmacophore model for STAT1/IRF7-dsDNA and OAS1/OAS3/IFIH1-dsRNA complexes, as well as for the genes binding to GDP or NADP+. The Lys amino acid was found commonly interacting with the ATP phosphate group from OAS1/EIF2AK2/IFIH1 genes. Considering the premise that targeting IRDS genes mediated resistance offers an efficient strategy to resensitize tumour cells and enhances the outcome of anti-cancer treatment, this review can add some novel insights to the field.
Keywords: ATP; DNA; DNA damage; IRDS genes; RNA; chemotherapy and radiotherapy; functional site; interferon; protein interfaces; receptors; resistance; upstream regulator; viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer.Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18490-5. doi: 10.1073/pnas.0809242105. Epub 2008 Nov 10. Proc Natl Acad Sci U S A. 2008. PMID: 19001271 Free PMC article.
-
Targeting interferon response genes sensitizes aromatase inhibitor resistant breast cancer cells to estrogen-induced cell death.Breast Cancer Res. 2015 Jan 15;17(1):6. doi: 10.1186/s13058-014-0506-7. Breast Cancer Res. 2015. PMID: 25588716 Free PMC article.
-
Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3.J Biol Chem. 1995 Aug 18;270(33):19624-9. doi: 10.1074/jbc.270.33.19624. J Biol Chem. 1995. PMID: 7642650
-
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
-
Signal transduction in the interferon system.Semin Oncol. 1998 Feb;25(1 Suppl 1):14-22. Semin Oncol. 1998. PMID: 9482536 Review.
Cited by
-
Targeting MUC1-C Suppresses Chronic Activation of Cytosolic Nucleotide Receptors and STING in Triple-Negative Breast Cancer.Cancers (Basel). 2022 May 24;14(11):2580. doi: 10.3390/cancers14112580. Cancers (Basel). 2022. PMID: 35681561 Free PMC article.
-
Intertumoral heterogeneity impacts oncolytic vesicular stomatitis virus efficacy in mouse pancreatic cancer cells.J Virol. 2023 Sep 28;97(9):e0100523. doi: 10.1128/jvi.01005-23. Epub 2023 Sep 6. J Virol. 2023. PMID: 37671865 Free PMC article.
-
Chronic exposure to Cytolethal Distending Toxin (CDT) promotes a cGAS-dependent type I interferon response.Cell Mol Life Sci. 2021 Sep;78(17-18):6319-6335. doi: 10.1007/s00018-021-03902-x. Epub 2021 Jul 25. Cell Mol Life Sci. 2021. PMID: 34308492 Free PMC article.
-
MUC1-C dictates neuroendocrine lineage specification in pancreatic ductal adenocarcinomas.Carcinogenesis. 2022 Feb 11;43(1):67-76. doi: 10.1093/carcin/bgab097. Carcinogenesis. 2022. PMID: 34657147 Free PMC article.
-
Structural determinants of peptide-dependent TAP1-TAP2 transit passage targeted by viral proteins and altered by cancer-associated mutations.Comput Struct Biotechnol J. 2021 Sep 9;19:5072-5091. doi: 10.1016/j.csbj.2021.09.006. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34589184 Free PMC article.
References
-
- Sarhan J., Liu B.C., Muendlein H.I., Weindel C.G., Smirnova I., Tang A.Y., Ilyukha V., Sorokin M., Buzdin A., Fitzgerald K.A., et al. Constitutive interferon signaling maintains critical threshold of MLKL expression to license necroptosis. Cell Death Differ. 2019;26:332–347. doi: 10.1038/s41418-018-0122-7. - DOI - PMC - PubMed
-
- Snyder A.G., Hubbard N.W., Messmer M.N., Kofman S.B., Hagan C.E., Orozco S.L., Chiang K., Daniels B.P., Baker D., Oberst A. Intra tumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci. Immunol. 2019;4:eaaw2004. doi: 10.1126/sciimmunol.aaw2004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

