Inosine in Biology and Disease
- PMID: 33921764
- PMCID: PMC8072771
- DOI: 10.3390/genes12040600
Inosine in Biology and Disease
Abstract
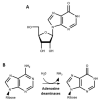
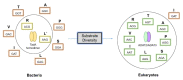
The nucleoside inosine plays an important role in purine biosynthesis, gene translation, and modulation of the fate of RNAs. The editing of adenosine to inosine is a widespread post-transcriptional modification in transfer RNAs (tRNAs) and messenger RNAs (mRNAs). At the wobble position of tRNA anticodons, inosine profoundly modifies codon recognition, while in mRNA, inosines can modify the sequence of the translated polypeptide or modulate the stability, localization, and splicing of transcripts. Inosine is also found in non-coding and exogenous RNAs, where it plays key structural and functional roles. In addition, molecular inosine is an important secondary metabolite in purine metabolism that also acts as a molecular messenger in cell signaling pathways. Here, we review the functional roles of inosine in biology and their connections to human health.
Keywords: RNA modification; adenosine deaminase acting on RNAs; deamination; inosine; translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Human tRNAs with inosine 34 are essential to efficiently translate eukarya-specific low-complexity proteins.Nucleic Acids Res. 2021 Jul 9;49(12):7011-7034. doi: 10.1093/nar/gkab461. Nucleic Acids Res. 2021. PMID: 34125917 Free PMC article.
-
Codon adaptation to tRNAs with Inosine modification at position 34 is widespread among Eukaryotes and present in two Bacterial phyla.RNA Biol. 2018;15(4-5):500-507. doi: 10.1080/15476286.2017.1358348. Epub 2017 Sep 26. RNA Biol. 2018. PMID: 28880718 Free PMC article.
-
A-to-I editing on tRNAs: biochemical, biological and evolutionary implications.FEBS Lett. 2014 Nov 28;588(23):4279-86. doi: 10.1016/j.febslet.2014.09.025. Epub 2014 Sep 27. FEBS Lett. 2014. PMID: 25263703 Review.
-
Celebrating wobble decoding: Half a century and still much is new.RNA Biol. 2018;15(4-5):537-553. doi: 10.1080/15476286.2017.1356562. Epub 2017 Sep 21. RNA Biol. 2018. PMID: 28812932 Free PMC article. Review.
-
Inosine RNA modifications are enriched at the codon wobble position in mouse oocytes and eggs†.Biol Reprod. 2019 Nov 21;101(5):938-949. doi: 10.1093/biolre/ioz130. Biol Reprod. 2019. PMID: 31346607 Free PMC article.
Cited by
-
SFMBT2 regulates plumage color via serum metabolites in Chinese Anyi tile-like gray chickens.Poult Sci. 2024 Dec;103(12):104391. doi: 10.1016/j.psj.2024.104391. Epub 2024 Oct 9. Poult Sci. 2024. PMID: 39427420 Free PMC article.
-
Uric acid and alterations of purine recycling disorders in Parkinson's disease: a cross-sectional study.NPJ Parkinsons Dis. 2024 Sep 9;10(1):170. doi: 10.1038/s41531-024-00785-0. NPJ Parkinsons Dis. 2024. PMID: 39251680 Free PMC article.
-
Human tRNAs with inosine 34 are essential to efficiently translate eukarya-specific low-complexity proteins.Nucleic Acids Res. 2021 Jul 9;49(12):7011-7034. doi: 10.1093/nar/gkab461. Nucleic Acids Res. 2021. PMID: 34125917 Free PMC article.
-
The Fight against the Carcinogenic Epstein-Barr Virus: Gut Microbiota, Natural Medicines, and Beyond.Int J Mol Sci. 2023 Jan 15;24(2):1716. doi: 10.3390/ijms24021716. Int J Mol Sci. 2023. PMID: 36675232 Free PMC article. Review.
-
Modifications of the human tRNA anticodon loop and their associations with genetic diseases.Cell Mol Life Sci. 2021 Dec;78(23):7087-7105. doi: 10.1007/s00018-021-03948-x. Epub 2021 Oct 4. Cell Mol Life Sci. 2021. PMID: 34605973 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

