Direct Conjugation of NEDD8 to the N-Terminus of a Model Protein Can Induce Degradation
- PMID: 33918652
- PMCID: PMC8069691
- DOI: 10.3390/cells10040854
Direct Conjugation of NEDD8 to the N-Terminus of a Model Protein Can Induce Degradation
Abstract
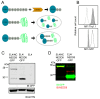
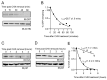
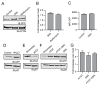
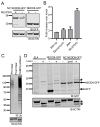
While the role of ubiquitin in protein degradation is well established, the role of other ubiquitin-like proteins (UBLs) in protein degradation is less clear. Neural precursor cell expressed developmentally down-regulated protein 8 (NEDD8) is the UBL with the highest level of amino acids identified when compared to ubiquitin. Here we tested if the N-terminal addition of NEDD8 to a protein of interest could lead to degradation. Mutation of critical glycine residues required for normal NEDD8 processing resulted in a non-cleavable fusion protein that was rapidly degraded within the cells by both the proteasome and autophagy. Both degradation pathways were dependent on a functional ubiquitin-conjugation system as treatment with MLN7243 increased levels of non-cleavable NEDD8-GFP. The degradation of non-cleavable, N-terminal NEDD8-GFP was not due to a failure of GFP folding as different NEDD8-GFP constructs with differing abilities to fold and fluoresce were similarly degraded. Though the fusion of NEDD8 to a protein resulted in degradation, treatment of cells with MLN4924, an inhibitor of the E1 activating enzyme for NEDD8, failed to prevent degradation of other destabilized substrates. Taken together these data suggest that under certain conditions, such as the model system described here, the covalent linkage of NEDD8 to a protein substrate may result in the target proteins degradation.
Keywords: NEDD8; proteasome; protein degradation; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
MLN4924 Inhibits Defective Ribosomal Product Antigen Presentation Independently of Direct NEDDylation of Protein Antigens.J Immunol. 2022 May 15;208(10):2273-2282. doi: 10.4049/jimmunol.2100584. Epub 2022 Apr 15. J Immunol. 2022. PMID: 35428693 Free PMC article.
-
Suppression of autophagy during mitosis via CUL4-RING ubiquitin ligases-mediated WIPI2 polyubiquitination and proteasomal degradation.Autophagy. 2019 Nov;15(11):1917-1934. doi: 10.1080/15548627.2019.1596484. Epub 2019 Mar 30. Autophagy. 2019. PMID: 30898011 Free PMC article.
-
Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes.Biochem J. 2012 Feb 1;441(3):927-36. doi: 10.1042/BJ20111671. Biochem J. 2012. PMID: 22004789 Free PMC article.
-
The Many Potential Fates of Non-Canonical Protein Substrates Subject to NEDDylation.Cells. 2021 Oct 5;10(10):2660. doi: 10.3390/cells10102660. Cells. 2021. PMID: 34685640 Free PMC article. Review.
-
The role of allostery in the ubiquitin-proteasome system.Crit Rev Biochem Mol Biol. 2013 Mar-Apr;48(2):89-97. doi: 10.3109/10409238.2012.742856. Epub 2012 Dec 13. Crit Rev Biochem Mol Biol. 2013. PMID: 23234564 Free PMC article. Review.
Cited by
-
Mechanisms and regulation of substrate degradation by the 26S proteasome.Nat Rev Mol Cell Biol. 2024 Oct 3. doi: 10.1038/s41580-024-00778-0. Online ahead of print. Nat Rev Mol Cell Biol. 2024. PMID: 39362999 Review.
-
UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas Plecoglossicida in Large Yellow Croaker (Larimichthys crocea).Int J Mol Sci. 2022 Jul 27;23(15):8298. doi: 10.3390/ijms23158298. Int J Mol Sci. 2022. PMID: 35955424 Free PMC article.
-
Cellular stress increases DRIP production and MHC Class I antigen presentation.Front Immunol. 2024 Aug 23;15:1445338. doi: 10.3389/fimmu.2024.1445338. eCollection 2024. Front Immunol. 2024. PMID: 39247192 Free PMC article.
-
Fast friends - Ubiquitin-like modifiers as engineered fusion partners.Semin Cell Dev Biol. 2022 Dec;132:132-145. doi: 10.1016/j.semcdb.2021.11.013. Epub 2021 Nov 25. Semin Cell Dev Biol. 2022. PMID: 34840080 Free PMC article. Review.
-
Variations in Cell Surface ACE2 Levels Alter Direct Binding of SARS-CoV-2 Spike Protein and Viral Infectivity: Implications for Measuring Spike Protein Interactions with Animal ACE2 Orthologs.J Virol. 2022 Sep 14;96(17):e0025622. doi: 10.1128/jvi.00256-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000847 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

