Information Encoded by the Flavivirus Genomes beyond the Nucleotide Sequence
- PMID: 33916729
- PMCID: PMC8038387
- DOI: 10.3390/ijms22073738
Information Encoded by the Flavivirus Genomes beyond the Nucleotide Sequence
Abstract
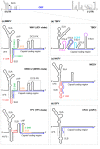
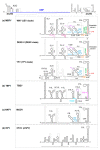
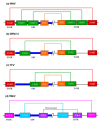
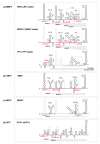
The genus Flavivirus comprises numerous, small, single positive-stranded RNA viruses, many of which are important human pathogens. To store all the information required for their successful propagation, flaviviruses use discrete structural genomic RNA elements to code for functional information by the establishment of dynamic networks of long-range RNA-RNA interactions that promote specific folding. These structural elements behave as true cis-acting, non-coding RNAs (ncRNAs) and have essential regulatory roles in the viral cycle. These include the control of the formation of subgenomic RNAs, known as sfRNAs, via the prevention of the complete degradation of the RNA genome. These sfRNAs are important in ensuring viral fitness. This work summarizes our current knowledge of the functions performed by the genome conformations and the role of RNA-RNA interactions in these functions. It also reviews the role of RNA structure in the production of sfRNAs across the genus Flavivirus, and their existence in related viruses.
Keywords: Flavivirus; RNA genomes; interactome; long-distant RNA–RNA interactions; sfRNAs; structure/function of RNA domains.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Zika Virus Subgenomic Flavivirus RNA Generation Requires Cooperativity between Duplicated RNA Structures That Are Essential for Productive Infection in Human Cells.J Virol. 2020 Aug 31;94(18):e00343-20. doi: 10.1128/JVI.00343-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32581095 Free PMC article.
-
Short Direct Repeats in the 3' Untranslated Region Are Involved in Subgenomic Flaviviral RNA Production.J Virol. 2020 Feb 28;94(6):e01175-19. doi: 10.1128/JVI.01175-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31896596 Free PMC article.
-
Standing your ground to exoribonucleases: Function of Flavivirus long non-coding RNAs.Virus Res. 2016 Jan 2;212:70-7. doi: 10.1016/j.virusres.2015.09.009. Epub 2015 Sep 11. Virus Res. 2016. PMID: 26368052 Free PMC article. Review.
-
Subgenomic flaviviral RNAs: What do we know after the first decade of research.Antiviral Res. 2018 Nov;159:13-25. doi: 10.1016/j.antiviral.2018.09.006. Epub 2018 Sep 11. Antiviral Res. 2018. PMID: 30217649 Review.
-
RNA Structure Duplications and Flavivirus Host Adaptation.Trends Microbiol. 2016 Apr;24(4):270-283. doi: 10.1016/j.tim.2016.01.002. Epub 2016 Feb 3. Trends Microbiol. 2016. PMID: 26850219 Free PMC article. Review.
Cited by
-
Encephalitic Arboviruses of Africa: Emergence, Clinical Presentation and Neuropathogenesis.Front Immunol. 2021 Dec 23;12:769942. doi: 10.3389/fimmu.2021.769942. eCollection 2021. Front Immunol. 2021. PMID: 35003087 Free PMC article. Review.
-
A safer cell-based yellow fever live attenuated vaccine protects mice against YFV infection.iScience. 2024 Sep 17;27(10):110972. doi: 10.1016/j.isci.2024.110972. eCollection 2024 Oct 18. iScience. 2024. PMID: 39398246 Free PMC article.
-
Yellow Fever Outbreak in Eastern Senegal, 2020-2021.Viruses. 2021 Jul 28;13(8):1475. doi: 10.3390/v13081475. Viruses. 2021. PMID: 34452343 Free PMC article.
-
Antisense Oligonucleotide-Based Therapy of Viral Infections.Pharmaceutics. 2021 Nov 26;13(12):2015. doi: 10.3390/pharmaceutics13122015. Pharmaceutics. 2021. PMID: 34959297 Free PMC article. Review.
-
Inter- and Intramolecular RNA-RNA Interactions Modulate the Regulation of Translation Mediated by the 3' UTR in West Nile Virus.Int J Mol Sci. 2023 Mar 10;24(6):5337. doi: 10.3390/ijms24065337. Int J Mol Sci. 2023. PMID: 36982407 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

