On the Origin of SARS-CoV-2: Did Cell Culture Experiments Lead to Increased Virulence of the Progenitor Virus for Humans?
- PMID: 33910809
- PMCID: PMC8193286
- DOI: 10.21873/invivo.12384
On the Origin of SARS-CoV-2: Did Cell Culture Experiments Lead to Increased Virulence of the Progenitor Virus for Humans?
Abstract
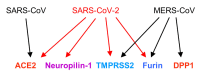
We are currently in a rapidly expanding pandemic of the SARS-CoV-2 virus, which originated in the city of Wuhan in central China. The disease COVID-19 is now spread worldwide and has tremendous socio-economic consequences. The origin of the virus can be reconstructed through epidemiological studies and, even more so, from genome comparisons. How the evolution of the virus and the transition to humans might have happened is the subject of much speculation. It is considered certain that the virus is of animal origin and very likely passed from bats to humans in a zoonotic event. An intermediate host was postulated, but many SARS-like bat viruses have the ability to infect human cells directly, which has been shown experimentally by scientists in the Wuhan Institute of Virology using collected specimens containing virus material from horseshoe bats. The propagation of SARS-like bat viruses in cell culture allowed experiments aimed at increasing the infectivity of the virus and adaptation to human cells. This article summarizes the unique properties of SARS-CoV-2 and focusses on a specific sequence encoding the spike protein. Possible scenarios of virus evolution are discussed, with particular emphasis on the hypothesis that the virus could have emerged unintentionally through routine culture or gain-of-function experiments in a laboratory, where it was optimally adapted to human cells and caused cryptic infections among workers who finally spread the virus causing the pandemic.
Keywords: Corona virus; Covid-19; SARS-CoV-2; bat viruses; gain-of-function experiments; pandemic; review.
Copyright© 2021, International Institute of Anticancer Research (Dr. George J. Delinasios), All rights reserved.
Conflict of interest statement
The Author declares that there are no conflicts of interest related to this work.
Figures

Similar articles
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
-
Gain-of-function and origin of Covid19.Presse Med. 2023 Mar;52(1):104167. doi: 10.1016/j.lpm.2023.104167. Epub 2023 Jun 2. Presse Med. 2023. PMID: 37269978 Free PMC article.
-
Genomic characterization and phylogenetic evolution of the SARS-CoV-2.Acta Virol. 2020;64(4):496-500. doi: 10.4149/av_2020_403. Acta Virol. 2020. PMID: 32985209
-
SARS-CoV-2: Zoonotic origin of pandemic coronavirus.Acta Virol. 2020;64(3):281-287. doi: 10.4149/av_2020_302. Acta Virol. 2020. PMID: 32985202
-
What do we know about the origin of COVID-19 three years later?Rev Clin Esp (Barc). 2023 Apr;223(4):240-243. doi: 10.1016/j.rceng.2023.02.010. Epub 2023 Mar 16. Rev Clin Esp (Barc). 2023. PMID: 36933695 Free PMC article. Review.
Cited by
-
Emergence, evolution, and vaccine production approaches of SARS-CoV-2 virus: Benefits of getting vaccinated and common questions.Saudi J Biol Sci. 2022 Apr;29(4):1981-1997. doi: 10.1016/j.sjbs.2021.12.020. Epub 2021 Dec 13. Saudi J Biol Sci. 2022. PMID: 34924802 Free PMC article. Review.
-
Portable and Air Conditioner-Based Bio-Protection Devices to Prevent Airborne Infections in Acute and Long-Term Healthcare Facilities, Public Gathering Places, Public Transportation, and Similar Entities.Cureus. 2024 Mar 11;16(3):e55950. doi: 10.7759/cureus.55950. eCollection 2024 Mar. Cureus. 2024. PMID: 38469370 Free PMC article.
-
What we know and what we need to know about the origin of SARS-CoV-2.Environ Res. 2021 Sep;200:111785. doi: 10.1016/j.envres.2021.111785. Epub 2021 Jul 28. Environ Res. 2021. PMID: 34329631 Free PMC article.
-
There is no "origin" to SARS-CoV-2.Environ Res. 2022 May 1;207:112173. doi: 10.1016/j.envres.2021.112173. Epub 2021 Oct 6. Environ Res. 2022. PMID: 34626592 Free PMC article.
-
Why Are Physicists Involved in the Studies on the Origin of SARS-CoV-2?J Biomed Phys Eng. 2021 Aug 1;11(4):413-414. doi: 10.31661/jbpe.v0i0.2106-1361. eCollection 2021 Aug. J Biomed Phys Eng. 2021. PMID: 34458188 Free PMC article. No abstract available.
References
-
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
-
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
