Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA
- PMID: 33904405
- PMCID: PMC8116057
- DOI: 10.7554/eLife.65537
Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA
Abstract
Genes involved in disease resistance are some of the fastest evolving and most diverse components of genomes. Large numbers of nucleotide-binding, leucine-rich repeat (NLR) genes are found in plant genomes and are required for disease resistance. However, NLRs can trigger autoimmunity, disrupt beneficial microbiota or reduce fitness. It is therefore crucial to understand how NLRs are controlled. Here, we show that the RNA-binding protein FPA mediates widespread premature cleavage and polyadenylation of NLR transcripts, thereby controlling their functional expression and impacting immunity. Using long-read Nanopore direct RNA sequencing, we resolved the complexity of NLR transcript processing and gene annotation. Our results uncover a co-transcriptional layer of NLR control with implications for understanding the regulatory and evolutionary dynamics of NLRs in the immune responses of plants.
Keywords: A. thaliana; NLR; alternative polyadenylation; chromosomes; direct RNA sequencing; gene expression; m6A; nanopore; plant biology; spen.
© 2021, Parker et al.
Conflict of interest statement
MP, KK, VZ, AS, DT, XY, PM, JB, SM, GB, GS No competing interests declared
Figures



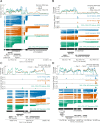
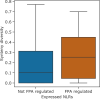
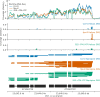
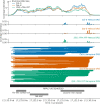



Similar articles
-
A dominant-interfering camta3 mutation compromises primary transcriptional outputs mediated by both cell surface and intracellular immune receptors in Arabidopsis thaliana.New Phytol. 2018 Mar;217(4):1667-1680. doi: 10.1111/nph.14943. Epub 2017 Dec 11. New Phytol. 2018. PMID: 29226970 Free PMC article.
-
The spen family protein FPA controls alternative cleavage and polyadenylation of RNA.Dev Cell. 2010 Feb 16;18(2):203-13. doi: 10.1016/j.devcel.2009.12.009. Epub 2010 Jan 14. Dev Cell. 2010. PMID: 20079695
-
Transcription termination and chimeric RNA formation controlled by Arabidopsis thaliana FPA.PLoS Genet. 2013 Oct;9(10):e1003867. doi: 10.1371/journal.pgen.1003867. Epub 2013 Oct 31. PLoS Genet. 2013. PMID: 24204292 Free PMC article.
-
Transcript-level expression control of plant NLR genes.Mol Plant Pathol. 2018 May;19(5):1267-1281. doi: 10.1111/mpp.12607. Epub 2017 Nov 20. Mol Plant Pathol. 2018. PMID: 28834153 Free PMC article. Review.
-
Advancement of research on plant NLRs evolution, biochemical activity, structural association, and engineering.Planta. 2020 Nov 12;252(6):101. doi: 10.1007/s00425-020-03512-0. Planta. 2020. PMID: 33180185 Review.
Cited by
-
CRISPR/dCas13(Rx) Derived RNA N6-methyladenosine (m6A) Dynamic Modification in Plant.Adv Sci (Weinh). 2024 Oct;11(39):e2401118. doi: 10.1002/advs.202401118. Epub 2024 Sep 4. Adv Sci (Weinh). 2024. PMID: 39229923 Free PMC article.
-
Catalytic activities, molecular connections, and biological functions of plant RNA exosome complexes.Plant Cell. 2022 Mar 4;34(3):967-988. doi: 10.1093/plcell/koab310. Plant Cell. 2022. PMID: 34954803 Free PMC article. Review.
-
Prion-like Proteins in Plants: Key Regulators of Development and Environmental Adaptation via Phase Separation.Plants (Basel). 2024 Sep 23;13(18):2666. doi: 10.3390/plants13182666. Plants (Basel). 2024. PMID: 39339640 Free PMC article. Review.
-
Long-read direct RNA sequencing reveals epigenetic regulation of chimeric gene-transposon transcripts in Arabidopsis thaliana.Nat Commun. 2023 Jun 5;14(1):3248. doi: 10.1038/s41467-023-38954-z. Nat Commun. 2023. PMID: 37277361 Free PMC article.
-
Two zinc finger proteins with functions in m6A writing interact with HAKAI.Nat Commun. 2022 Mar 2;13(1):1127. doi: 10.1038/s41467-022-28753-3. Nat Commun. 2022. PMID: 35236848 Free PMC article.
References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM075060/GM/NIGMS NIH HHS/United States
- BB/M010066/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J00247X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M004155/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

