Myosin 7b is a regulatory long noncoding RNA (lncMYH7b) in the human heart
- PMID: 33895132
- PMCID: PMC8141895
- DOI: 10.1016/j.jbc.2021.100694
Myosin 7b is a regulatory long noncoding RNA (lncMYH7b) in the human heart
Abstract
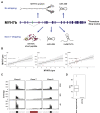
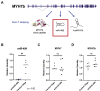
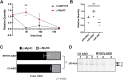
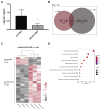
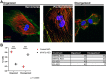
Myosin heavy chain 7b (MYH7b) is an ancient member of the myosin heavy chain motor protein family that is expressed in striated muscles. In mammalian cardiac muscle, MYH7b RNA is expressed along with two other myosin heavy chains, β-myosin heavy chain (β-MyHC) and α-myosin heavy chain (α-MyHC). However, unlike β-MyHC and α-MyHC, which are maintained in a careful balance at the protein level, the MYH7b locus does not produce a full-length protein in the heart due to a posttranscriptional exon-skipping mechanism that occurs in a tissue-specific manner. Whether this locus has a role in the heart beyond producing its intronic microRNA, miR-499, was unclear. Using cardiomyocytes derived from human induced pluripotent stem cells as a model system, we found that the noncoding exon-skipped RNA (lncMYH7b) affects the transcriptional landscape of human cardiomyocytes, independent of miR-499. Specifically, lncMYH7b regulates the ratio of β-MyHC to α-MyHC, which is crucial for cardiac contractility. We also found that lncMYH7b regulates beat rate and sarcomere formation in cardiomyocytes. This regulation is likely achieved through control of a member of the TEA domain transcription factor family (TEAD3, which is known to regulate β-MyHC). Therefore, we conclude that this ancient gene has been repurposed by alternative splicing to produce a regulatory long-noncoding RNA in the human heart that affects cardiac myosin composition.
Keywords: alternative splicing; gene regulation; heart failure; long noncoding RNA (lncRNA); myosin heavy chain (MyHC); myosin heavy chain 7b (MYH7b).
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest No conflicts of interest were reported.
Figures


Similar articles
-
Nonproductive Splicing Prevents Expression of MYH7b Protein in the Mammalian Heart.J Am Heart Assoc. 2021 Jul 20;10(14):e020965. doi: 10.1161/JAHA.121.020965. Epub 2021 Jul 6. J Am Heart Assoc. 2021. PMID: 34227390 Free PMC article.
-
Functional divergence of the sarcomeric myosin, MYH7b, supports species-specific biological roles.J Biol Chem. 2023 Jan;299(1):102657. doi: 10.1016/j.jbc.2022.102657. Epub 2022 Nov 9. J Biol Chem. 2023. PMID: 36334627 Free PMC article.
-
Calcium-independent negative inotropy by beta-myosin heavy chain gene transfer in cardiac myocytes.Circ Res. 2007 Apr 27;100(8):1182-90. doi: 10.1161/01.RES.0000264102.00706.4e. Epub 2007 Mar 15. Circ Res. 2007. PMID: 17363698
-
The ancient sarcomeric myosins found in specialized muscles.Skelet Muscle. 2019 Mar 5;9(1):7. doi: 10.1186/s13395-019-0192-3. Skelet Muscle. 2019. PMID: 30836986 Free PMC article. Review.
-
Structural implications of β-cardiac myosin heavy chain mutations in human disease.Anat Rec (Hoboken). 2014 Sep;297(9):1670-80. doi: 10.1002/ar.22973. Anat Rec (Hoboken). 2014. PMID: 25125180 Review.
Cited by
-
Cardiac Development Long Non-Coding RNA (CARDEL) Is Activated during Human Heart Development and Contributes to Cardiac Specification and Homeostasis.Cells. 2024 Jun 18;13(12):1050. doi: 10.3390/cells13121050. Cells. 2024. PMID: 38920678 Free PMC article.
-
Post-ischemic cardioprotective potential of exogenous ubiquitin in myocardial remodeling late after ischemia/reperfusion injury.Life Sci. 2023 Jan 1;312:121216. doi: 10.1016/j.lfs.2022.121216. Epub 2022 Nov 24. Life Sci. 2023. PMID: 36435225 Free PMC article.
-
Circulating miR-499a-5p Is a Potential Biomarker of MYH7-Associated Hypertrophic Cardiomyopathy.Int J Mol Sci. 2022 Mar 30;23(7):3791. doi: 10.3390/ijms23073791. Int J Mol Sci. 2022. PMID: 35409153 Free PMC article.
-
Long non-coding RNAs in cardiac hypertrophy and heart failure: functions, mechanisms and clinical prospects.Nat Rev Cardiol. 2024 May;21(5):326-345. doi: 10.1038/s41569-023-00952-5. Epub 2023 Nov 20. Nat Rev Cardiol. 2024. PMID: 37985696 Free PMC article. Review.
-
The status of the human gene catalogue.Nature. 2023 Oct;622(7981):41-47. doi: 10.1038/s41586-023-06490-x. Epub 2023 Oct 4. Nature. 2023. PMID: 37794265 Free PMC article. Review.
References
-
- Weiss A., Leinwand L. The mammalian myosin heavy chain gene family. Annu. Rev. Cell Dev. Biol. 1996;12:417–439. - PubMed
-
- Miyata S., Minobe W., Bristow M.R., Leinwand L.A. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ. Res. 2000;86:386–390. - PubMed
-
- Lowes B.D., Gilbert E.M., Abraham W.T., Minobe W.A., Larrabee P., Ferguson D., Wolfel E.E., Lindenfeld J.A., Tsvetkova T., Robertson A.D., Quaife R.A., Bristow M.R. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N. Engl. J. Med. 2002;346:1357–1365. - PubMed
-
- Hasegawa K., Lee S.J., Jobe S.M., Markham B.E., Kitsis R.N. cis-Acting sequences that mediate induction of beta-myosin heavy chain gene expression during left ventricular hypertrophy due to aortic constriction. Circulation. 1997;96:3943–3953. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

