High expression of COMMD7 is an adverse prognostic factor in acute myeloid leukemia
- PMID: 33891561
- PMCID: PMC8109082
- DOI: 10.18632/aging.202901
High expression of COMMD7 is an adverse prognostic factor in acute myeloid leukemia
Abstract
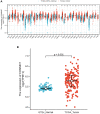
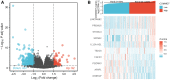
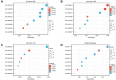
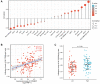
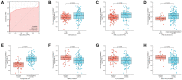
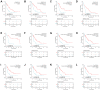
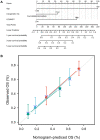
Acute myeloid leukemia (AML) is a frequent malignancy in adults worldwide; identifying preferable biomarkers has become one of the current challenges. Given that COMMD7 has been reported associated with tumor progression in various human solid cancers but rarely reported in AML, herein, RNA sequencing data from TCGA and GTEx were obtained for analysis of COMMD7 expression and differentially expressed gene (DEG). Furthermore, functional enrichment analysis of COMMD7-related DEGs was performed by GO/KEGG, GSEA, immune cell infiltration analysis, and protein-protein interaction (PPI) network. In addition, the clinical significance of COMMD7 in AML was figured out by Kaplan-Meier Cox regression and prognostic nomogram model. R package was used to analyze incorporated studies. As a result, COMMD7 was highly expressed in various malignancies, including AML, compared with normal samples. Moreover, high expression of COMMD7 was associated with poor prognosis in 151 AML samples, as well as subgroups with age >60, NPM1 mutation-positive, FLT3 mutation-negative, and DNMT3A mutation-negative, et al. (P < 0.05). High COMMD7 was an independent prognostic factor in Cox regression analysis; Age and cytogenetics risk were included in the nomogram prognostic model. Furthermore, a total of 529 DEGs were identified between the high- and the low- expression group, of which 92 genes were up-regulated and 437 genes were down-regulated. Collectively, high expression of COMMD7 is a potential biomarker for adverse outcomes in AML. The DEGs and pathways recognized in the study provide a preliminary grasp of the underlying molecular mechanisms of AML carcinogenesis and progression.
Keywords: COMMD7; R packages; The Cancer Genome Atlas; acute myeloid leukemia.
Conflict of interest statement
Figures


Similar articles
-
Bioinformatics Analysis Identifies Key Genes and Pathways in Acute Myeloid Leukemia Associated with DNMT3A Mutation.Biomed Res Int. 2020 Nov 23;2020:9321630. doi: 10.1155/2020/9321630. eCollection 2020. Biomed Res Int. 2020. PMID: 33299888 Free PMC article.
-
High expression of VTA1 is an adverse prognostic factor in lung adenocarcinoma.Cell Mol Biol (Noisy-le-grand). 2024 Jan 31;70(1):46-55. doi: 10.14715/cmb/2024.70.1.7. Cell Mol Biol (Noisy-le-grand). 2024. PMID: 38372114
-
Construction of prognostic risk prediction model based on high-throughput sequencing expression profile data in childhood acute myeloid leukemia.Blood Cells Mol Dis. 2019 Jul;77:43-50. doi: 10.1016/j.bcmd.2019.03.008. Epub 2019 Mar 28. Blood Cells Mol Dis. 2019. PMID: 30954792
-
A concise review on the molecular genetics of acute myeloid leukemia.Leuk Res. 2021 Dec;111:106727. doi: 10.1016/j.leukres.2021.106727. Epub 2021 Oct 16. Leuk Res. 2021. PMID: 34700049 Review.
-
CAPN1 is a novel biomaker of patients with AML based on comprehensive analysis.Biotechnol Genet Eng Rev. 2024 Dec;40(4):3884-3900. doi: 10.1080/02648725.2023.2204688. Epub 2023 Apr 28. Biotechnol Genet Eng Rev. 2024. PMID: 37114994
Cited by
-
BTG2 and SerpinB5, a novel gene pair to evaluate the prognosis of lung adenocarcinoma.Front Immunol. 2023 Mar 17;14:1098700. doi: 10.3389/fimmu.2023.1098700. eCollection 2023. Front Immunol. 2023. PMID: 37006240 Free PMC article.
-
Mining of transcriptome identifies CD109 and LRP12 as possible biomarkers and deregulation mechanism of T cell receptor pathway in Acute Myeloid Leukemia.Heliyon. 2022 Oct 18;8(10):e11123. doi: 10.1016/j.heliyon.2022.e11123. eCollection 2022 Oct. Heliyon. 2022. PMID: 36299526 Free PMC article.
-
Development of a novel pyroptosis-related LncRNA signature with multiple significance in acute myeloid leukemia.Front Genet. 2023 Jan 4;13:1029717. doi: 10.3389/fgene.2022.1029717. eCollection 2022. Front Genet. 2023. PMID: 36685973 Free PMC article.
-
ZNF460-regulated COMMD7 Promotes Acute Myeloid Leukemia Proliferation Via the NF-κB Signaling Pathway.Int J Med Sci. 2023 Feb 21;20(4):520-529. doi: 10.7150/ijms.80047. eCollection 2023. Int J Med Sci. 2023. PMID: 37057209 Free PMC article.
-
The Identification of APOBEC3G as a Potential Prognostic Biomarker in Acute Myeloid Leukemia and a Possible Drug Target for Crotonoside.Molecules. 2022 Sep 7;27(18):5804. doi: 10.3390/molecules27185804. Molecules. 2022. PMID: 36144542 Free PMC article.
References
-
- Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL, Lo-Coco F, Naoe T, et al.. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017; 129:424–47. 10.1182/blood-2016-08-733196 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

