Membrane Env Liposomes Facilitate Immunization with Multivalent Full-Length HIV Spikes
- PMID: 33883221
- PMCID: PMC8316072
- DOI: 10.1128/JVI.00005-21
Membrane Env Liposomes Facilitate Immunization with Multivalent Full-Length HIV Spikes
Abstract
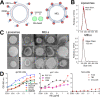
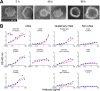
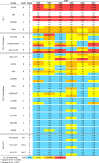
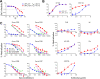
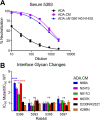
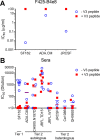
A major goal of HIV vaccine design is to elicit broadly neutralizing antibodies (bNAbs). Such bNAbs target HIV's trimeric, membrane-embedded envelope glycoprotein spikes (mEnv). Soluble Env (sEnv) trimers have been used as vaccines, but engineering sEnvs for stability, multivalency, and desired antigenicity is problematic and deletes key neutralizing epitopes on glycoprotein 41 (gp41) while creating neoepitopes that elicit unwanted antibodies. Meanwhile, multivalent mEnv vaccines are challenging to develop due to trimer instability and low mEnv copy number amid other extraneous proteins on virus-like particles. Here, we describe a multivalent mEnv vaccine platform that does not require protein engineering or extraneous proteins. mEnv trimers were fixed, purified, and combined with naked liposomes in mild detergent. On removal of detergent, mEnv spikes were observed embedded in liposome particles (mean diameter, 133 nm) in correct orientation. These particles were recognized by HIV bNAbs and not non-NAbs and are designated
Keywords: HIV; envelope; gp120; gp41; immunogen; liposome; vaccine.
Figures




Similar articles
-
Potent Induction of Envelope-Specific Antibody Responses by Virus-Like Particle Immunogens Based on HIV-1 Envelopes from Patients with Early Broadly Neutralizing Responses.J Virol. 2022 Jan 12;96(1):e0134321. doi: 10.1128/JVI.01343-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668778 Free PMC article.
-
A Trimeric HIV-1 Envelope gp120 Immunogen Induces Potent and Broad Anti-V1V2 Loop Antibodies against HIV-1 in Rabbits and Rhesus Macaques.J Virol. 2018 Feb 12;92(5):e01796-17. doi: 10.1128/JVI.01796-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237847 Free PMC article.
-
Immunogenic Display of Purified Chemically Cross-Linked HIV-1 Spikes.J Virol. 2015 Jul;89(13):6725-45. doi: 10.1128/JVI.03738-14. Epub 2015 Apr 15. J Virol. 2015. PMID: 25878116 Free PMC article.
-
GP120: target for neutralizing HIV-1 antibodies.Annu Rev Immunol. 2006;24:739-69. doi: 10.1146/annurev.immunol.24.021605.090557. Annu Rev Immunol. 2006. PMID: 16551265 Review.
-
Strategies for eliciting multiple lineages of broadly neutralizing antibodies to HIV by vaccination.Curr Opin Virol. 2021 Dec;51:172-178. doi: 10.1016/j.coviro.2021.09.015. Epub 2021 Nov 4. Curr Opin Virol. 2021. PMID: 34742037 Free PMC article. Review.
Cited by
-
Functional Delineation of a Protein-Membrane Interaction Hotspot Site on the HIV-1 Neutralizing Antibody 10E8.Int J Mol Sci. 2022 Sep 15;23(18):10767. doi: 10.3390/ijms231810767. Int J Mol Sci. 2022. PMID: 36142694 Free PMC article.
References
-
- UNAIDS. 2020. UNAIDS data 2020. https://www.unaids.org/sites/default/files/media_asset/2020_aids-data-bo.... Accessed 5 Nov 2020.
-
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. 10.1056/NEJMoa0908492. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

