Diagnostic and therapeutic pitfalls in NPM1-mutated AML: notes from the field
- PMID: 33879827
- PMCID: PMC8056374
- DOI: 10.1038/s41375-021-01222-4
Diagnostic and therapeutic pitfalls in NPM1-mutated AML: notes from the field
Abstract
Mutations of Nucleophosmin (NPM1) are the most common genetic abnormalities in adult acute myeloid leukaemia (AML), accounting for about 30% of cases. NPM1-mutated AML has been recognized as distinct entity in the 2017 World Health Organization (WHO) classification of lympho-haematopoietic neoplasms. WHO criteria allow recognition of this leukaemia entity and its distinction from AML with myelodysplasia-related changes, AML with BCR-ABL1 rearrangement and AML with RUNX1 mutations. Nevertheless, controversial issues include the percentage of blasts required for the diagnosis of NPM1-mutated AML and whether cases of NPM1-mutated myelodysplasia and chronic myelomonocytic leukaemia do exist. Evaluation of NPM1 and FLT3 status represents a major pillar of the European LeukemiaNet (ELN) genetic-based risk stratification model. Moreover, NPM1 mutations are particularly suitable for assessing measurable residual disease (MRD) since they are frequent, stable at relapse and do not drive clonal haematopoiesis. Ideally, combining monitoring of MRD with the ELN prognostication model can help to guide therapeutic decisions. Here, we provide examples of instructive cases of NPM1-mutated AML, in order to provide criteria for the appropriate diagnosis and therapy of this frequent leukaemia entity.
© 2021. The Author(s).
Conflict of interest statement
BF licensed a patent on
Figures
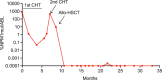
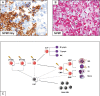
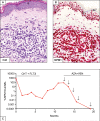
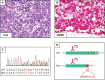
Similar articles
-
How I diagnose and treat NPM1-mutated AML.Blood. 2021 Feb 4;137(5):589-599. doi: 10.1182/blood.2020008211. Blood. 2021. PMID: 33171486
-
Criteria for Diagnosis and Molecular Monitoring of NPM1-Mutated AML.Blood Cancer Discov. 2024 Jan 8;5(1):8-20. doi: 10.1158/2643-3230.BCD-23-0144. Blood Cancer Discov. 2024. PMID: 37917833 Free PMC article. Review.
-
Measurable residual disease detected by flow cytometry independently predicts prognoses of NPM1-mutated acute myeloid leukemia.Ann Hematol. 2023 Feb;102(2):337-347. doi: 10.1007/s00277-022-05033-0. Epub 2022 Nov 15. Ann Hematol. 2023. PMID: 36378304
-
Minimal/Measurable Residual Disease Monitoring in NPM1-Mutated Acute Myeloid Leukemia: A Clinical Viewpoint and Perspectives.Int J Mol Sci. 2018 Nov 6;19(11):3492. doi: 10.3390/ijms19113492. Int J Mol Sci. 2018. PMID: 30404199 Free PMC article. Review.
-
Validation of the 2017 European LeukemiaNet classification for acute myeloid leukemia with NPM1 and FLT3-internal tandem duplication genotypes.Cancer. 2019 Apr 1;125(7):1091-1100. doi: 10.1002/cncr.31885. Epub 2018 Dec 6. Cancer. 2019. PMID: 30521114 Free PMC article.
Cited by
-
Liquid-liquid phase separation in tumor biology.Signal Transduct Target Ther. 2022 Jul 8;7(1):221. doi: 10.1038/s41392-022-01076-x. Signal Transduct Target Ther. 2022. PMID: 35803926 Free PMC article. Review.
-
Gilteritinib combination therapies in pediatric patients with FLT3-mutated acute myeloid leukemia.Blood Adv. 2021 Dec 14;5(23):5215-5219. doi: 10.1182/bloodadvances.2021005164. Blood Adv. 2021. PMID: 34592761 Free PMC article. No abstract available.
-
The Impact of Mutation of Myelodysplasia-Related Genes in De Novo Acute Myeloid Leukemia Carrying NPM1 Mutation.Cancers (Basel). 2022 Dec 29;15(1):198. doi: 10.3390/cancers15010198. Cancers (Basel). 2022. PMID: 36612194 Free PMC article.
-
Super-Enhancers, Phase-Separated Condensates, and 3D Genome Organization in Cancer.Cancers (Basel). 2022 Jun 10;14(12):2866. doi: 10.3390/cancers14122866. Cancers (Basel). 2022. PMID: 35740532 Free PMC article. Review.
-
Biomolecular condensates: Formation mechanisms, biological functions, and therapeutic targets.MedComm (2020). 2023 Feb 28;4(2):e223. doi: 10.1002/mco2.223. eCollection 2023 Apr. MedComm (2020). 2023. PMID: 36875159 Free PMC article. Review.
References
-
- Arber DA, Brunning RD, Le Beau MM, Falini B, Vardiman JW, Porwit A, et al. Acute myeloid leukaemia with recurrent genetic abnormalities. In: Swerdlow S et al. editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer (IARC); 2017. p. 130–49.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

