Multifactorial Traits of SARS-CoV-2 Cell Entry Related to Diverse Host Proteases and Proteins
- PMID: 33875625
- PMCID: PMC8094071
- DOI: 10.4062/biomolther.2021.048
Multifactorial Traits of SARS-CoV-2 Cell Entry Related to Diverse Host Proteases and Proteins
Abstract
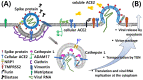
The most effective way to control newly emerging infectious disease, such as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, is to strengthen preventative or therapeutic public health strategies before the infection spreads worldwide. However, global health systems remain at the early stages in anticipating effective therapeutics or vaccines to combat the SARS-CoV-2 pandemic. While maintaining social distance is the most crucial metric to avoid spreading the virus, symptomatic therapy given to patients on the clinical manifestations helps save lives. The molecular properties of SARS-CoV-2 infection have been quickly elucidated, paving the way to therapeutics, vaccine development, and other medical interventions. Despite this progress, the detailed biomolecular mechanism of SARS-CoV-2 infection remains elusive. Given virus invasion of cells is a determining factor for virulence, understanding the viral entry process can be a mainstay in controlling newly emerged viruses. Since viral entry is mediated by selective cellular proteases or proteins associated with receptors, identification and functional analysis of these proteins could provide a way to disrupt virus propagation. This review comprehensively discusses cellular machinery necessary for SARS-CoV-2 infection. Understanding multifactorial traits of the virus entry will provide a substantial guide to facilitate antiviral drug development.
Keywords: Antiviral drugs; Cell entry; Cellular proteins; SARS-CoV-2.
Figures

Similar articles
-
Molecular Insights of SARS-CoV-2 Infection and Molecular Treatments.Curr Mol Med. 2022;22(7):621-639. doi: 10.2174/1566524021666211013121831. Curr Mol Med. 2022. PMID: 34645374 Review.
-
Therapeutic potential of green tea catechin, (-)-epigallocatechin-3-O-gallate (EGCG) in SARS-CoV-2 infection: Major interactions with host/virus proteases.Phytomed Plus. 2023 Feb;3(1):100402. doi: 10.1016/j.phyplu.2022.100402. Epub 2022 Dec 30. Phytomed Plus. 2023. PMID: 36597465 Free PMC article. Review.
-
Host Cell Proteases Mediating SARS-CoV-2 Entry: An Overview.Curr Top Med Chem. 2022;22(21):1776-1792. doi: 10.2174/1568026622666220726122339. Curr Top Med Chem. 2022. PMID: 35894476 Review.
-
Structural understanding of SARS-CoV-2 virus entry to host cells.Front Mol Biosci. 2023 Nov 2;10:1288686. doi: 10.3389/fmolb.2023.1288686. eCollection 2023. Front Mol Biosci. 2023. PMID: 38033388 Free PMC article. Review.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
Cited by
-
A Closer Look at ACE2 Signaling Pathway and Processing during COVID-19 Infection: Identifying Possible Targets.Vaccines (Basel). 2022 Dec 21;11(1):13. doi: 10.3390/vaccines11010013. Vaccines (Basel). 2022. PMID: 36679858 Free PMC article. Review.
References
-
- Ami Y., Nagata N., Shirato K., Watanabe R., Iwata N., Nakagaki K., Fukushi S., Saijo M., Morikawa S., Taguchi F. Co-infection of respiratory bacterium with severe acute respiratory syndrome coronavirus induces an exacerbated pneumonia in mice. Microbiol. Immunol. 2008;52:118–127. doi: 10.1111/j.1348-0421.2008.00011.x. - DOI - PMC - PubMed
-
- Baron J., Tarnow C., Mayoli-Nüssle D., Schilling E., Meyer D., Hammami M., Schwalm F., Steinmetzer T., Guan Y., Garten W., Klenk H. D., Böttcher-Friebertshäuser E. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J. Virol. 2013;87:1811–1820. doi: 10.1128/JVI.02320-12. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

