Inclusion Membrane Growth and Composition Are Altered by Overexpression of Specific Inclusion Membrane Proteins in Chlamydia trachomatis L2
- PMID: 33875478
- PMCID: PMC8208519
- DOI: 10.1128/IAI.00094-21
Inclusion Membrane Growth and Composition Are Altered by Overexpression of Specific Inclusion Membrane Proteins in Chlamydia trachomatis L2
Abstract
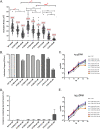
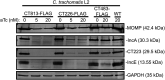
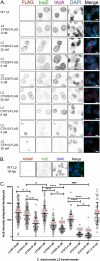
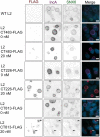
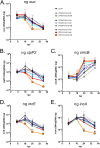
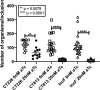
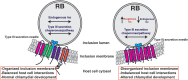
Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections. This obligate intracellular bacterium develops within a membrane-bound vacuole called an inclusion, which sequesters the chlamydiae from the host cytoplasm. Host-pathogen interactions at this interface are mediated by chlamydial inclusion membrane proteins (Incs). However, the specific functions of most Incs are poorly characterized. Previous work from our laboratories indicated that expressing an IncF fusion protein at high levels in C. trachomatis L2 negatively impacted inclusion expansion and progeny production. We hypothesize that some Incs function in the structure and organization of the inclusion membrane and that overexpression of those Incs will alter the composition of endogenous Incs within the inclusion membrane. Consequently, inclusion biogenesis and chlamydial development are negatively impacted. To investigate this, C. trachomatis L2 was transformed with inducible expression plasmids encoding IncF-, CT813-, or CT226-FLAG. Overexpression of IncF-FLAG or CT813-FLAG, but not CT226-FLAG, altered chlamydial development, as demonstrated by smaller inclusions, fewer progeny, and increased plasmid loss. The overexpression of CT813-FLAG reduced the detectable levels of endogenous IncE and IncG in the inclusion membrane. Notably, recruitment of sorting nexin-6, a eukaryotic protein binding partner of IncE, was also reduced after CT813 overexpression. Gene expression studies and ultrastructural analysis of chlamydial organisms demonstrated that chlamydial development was altered when CT813-FLAG was overexpressed. Overall, these data indicate that disrupting the expression of specific Incs changed the composition of Incs within the inclusion membrane and the recruitment of associated host cell proteins, which negatively impacted C. trachomatis development.
Keywords: Chlamydia trachomatis; Inc; chlamydia; developmental cycle; inclusion membrane; type III secretion.
Figures
Similar articles
-
Chlamydia trachomatis Inc Ct226 is vital for FLI1 and LRRF1 recruitment to the chlamydial inclusion.mSphere. 2024 Nov 21;9(11):e0047324. doi: 10.1128/msphere.00473-24. Epub 2024 Oct 15. mSphere. 2024. PMID: 39404459 Free PMC article.
-
Eukaryotic SNARE VAMP3 Dynamically Interacts with Multiple Chlamydial Inclusion Membrane Proteins.Infect Immun. 2021 Jan 19;89(2):e00409-20. doi: 10.1128/IAI.00409-20. Print 2021 Jan 19. Infect Immun. 2021. PMID: 33229367 Free PMC article.
-
Development of a Proximity Labeling System to Map the Chlamydia trachomatis Inclusion Membrane.Front Cell Infect Microbiol. 2017 Feb 15;7:40. doi: 10.3389/fcimb.2017.00040. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28261569 Free PMC article.
-
Got mutants? How advances in chlamydial genetics have furthered the study of effector proteins.Pathog Dis. 2021 Feb 4;79(2):ftaa078. doi: 10.1093/femspd/ftaa078. Pathog Dis. 2021. PMID: 33512479 Free PMC article. Review.
-
[Effector proteins of Clamidia].Mol Biol (Mosk). 2009 Nov-Dec;43(6):963-83. Mol Biol (Mosk). 2009. PMID: 20088373 Review. Russian.
Cited by
-
Eukaryotic Clathrin Adapter Protein and Mediator of Cholesterol Homeostasis, PICALM, Affects Trafficking to the Chlamydial Inclusion.Mol Cell Biol. 2023 Feb 13;43(2):1-13. doi: 10.1080/10985549.2023.2171695. Online ahead of print. Mol Cell Biol. 2023. PMID: 36779337 Free PMC article.
-
Molecular pathogenesis of Chlamydia trachomatis.Front Cell Infect Microbiol. 2023 Oct 18;13:1281823. doi: 10.3389/fcimb.2023.1281823. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37920447 Free PMC article. Review.
-
Codon-Dependent Transcriptional Changes in Response to Tryptophan Limitation in the Tryptophan Auxotrophic Pathogens Chlamydia trachomatis and Streptococcus pyogenes.mSystems. 2021 Dec 21;6(6):e0126921. doi: 10.1128/mSystems.01269-21. Epub 2021 Dec 14. mSystems. 2021. PMID: 34904862 Free PMC article.
-
Host Cell Amplification of Nutritional Stress Contributes To Persistence in Chlamydia trachomatis.mBio. 2022 Dec 20;13(6):e0271922. doi: 10.1128/mbio.02719-22. Epub 2022 Nov 15. mBio. 2022. PMID: 36377897 Free PMC article.
-
The Chlamydia effector CpoS modulates the inclusion microenvironment and restricts the interferon response by acting on Rab35.mBio. 2023 Aug 31;14(4):e0319022. doi: 10.1128/mbio.03190-22. Epub 2023 Aug 2. mBio. 2023. PMID: 37530528 Free PMC article.
References
-
- CDC. 2017. Sexually transmitted disease surveillance 2017. CDC, Atlanta, GA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

