RbAp46/48LIN-53 and HAT-1 are required for initial CENP-AHCP-3 deposition and de novo holocentromere formation on artificial chromosomes in Caenorhabditis elegans embryos
- PMID: 33872374
- PMCID: PMC8450102
- DOI: 10.1093/nar/gkab217
RbAp46/48LIN-53 and HAT-1 are required for initial CENP-AHCP-3 deposition and de novo holocentromere formation on artificial chromosomes in Caenorhabditis elegans embryos
Abstract
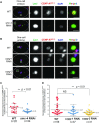
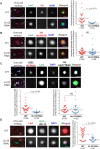
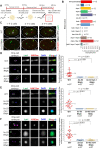
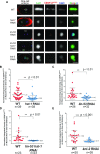
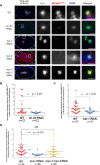
Foreign DNA microinjected into the Caenorhabditis elegans syncytial gonad forms episomal extra-chromosomal arrays, or artificial chromosomes (ACs), in embryos. Short, linear DNA fragments injected concatemerize into high molecular weight (HMW) DNA arrays that are visible as punctate DAPI-stained foci in oocytes, and they undergo chromatinization and centromerization in embryos. The inner centromere, inner kinetochore and spindle checkpoint components, including AIR-2, CENP-AHCP-3, Mis18BP1KNL-2 and BUB-1, respectively, assemble onto the nascent ACs during the first mitosis. The DNA replication efficiency of ACs improves over several cell cycles, which correlates with the improvement of kinetochore bi-orientation and proper segregation of ACs. Depletion of condensin II subunits, like CAPG-2 and SMC-4, but not the replicative helicase component, MCM-2, reduces de novo CENP-AHCP-3 level on nascent ACs. Furthermore, H3K9ac, H4K5ac and H4K12ac are highly enriched on newly chromatinized ACs. RbAp46/48LIN-53 and HAT-1, which affect the acetylation of histone H3 and H4, are essential for chromatinization, de novo centromere formation and segregation competency of nascent ACs. RbAp46/48LIN-53 or HAT-1 depletion causes the loss of both CENP-AHCP-3 and Mis18BP1KNL-2 initial deposition at de novo centromeres on ACs. This phenomenon is different from centromere maintenance on endogenous chromosomes, where Mis18BP1KNL-2 functions upstream of RbAp46/48LIN-53.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Formation of artificial chromosomes in Caenorhabditis elegans and analyses of their segregation in mitosis, DNA sequence composition and holocentromere organization.Nucleic Acids Res. 2021 Sep 20;49(16):9174-9193. doi: 10.1093/nar/gkab690. Nucleic Acids Res. 2021. PMID: 34417622 Free PMC article.
-
Histone H3K9 and H4 Acetylations and Transcription Facilitate the Initial CENP-AHCP-3 Deposition and De Novo Centromere Establishment in Caenorhabditis elegans Artificial Chromosomes.Epigenetics Chromatin. 2018 Apr 13;11(1):16. doi: 10.1186/s13072-018-0185-1. Epigenetics Chromatin. 2018. PMID: 29653589 Free PMC article.
-
RbAp46/48(LIN-53) Is Required for Holocentromere Assembly in Caenorhabditis elegans.Cell Rep. 2016 Mar 1;14(8):1819-28. doi: 10.1016/j.celrep.2016.01.065. Epub 2016 Feb 18. Cell Rep. 2016. PMID: 26904949
-
Construction and analysis of artificial chromosomes with de novo holocentromeres in Caenorhabditis elegans.Essays Biochem. 2020 Sep 4;64(2):233-249. doi: 10.1042/EBC20190067. Essays Biochem. 2020. PMID: 32756873 Review.
-
"Lessons from the extremes: Epigenetic and genetic regulation in point monocentromere and holocentromere establishment on artificial chromosomes".Exp Cell Res. 2020 May 15;390(2):111974. doi: 10.1016/j.yexcr.2020.111974. Epub 2020 Mar 26. Exp Cell Res. 2020. PMID: 32222413 Review.
Cited by
-
Mitotic chromosome condensation requires phosphorylation of the centromeric protein KNL-2 in C. elegans.J Cell Sci. 2021 Dec 1;134(23):jcs259088. doi: 10.1242/jcs.259088. Epub 2021 Dec 2. J Cell Sci. 2021. PMID: 34734636 Free PMC article.
-
Transgenerational inheritance of centromere identity requires the CENP-A N-terminal tail in the C. elegans maternal germ line.PLoS Biol. 2021 Jul 6;19(7):e3000968. doi: 10.1371/journal.pbio.3000968. eCollection 2021 Jul. PLoS Biol. 2021. PMID: 34228701 Free PMC article.
-
Argonaute protein CSR-1 restricts localization of holocentromere protein HCP-3, the C. elegans CENP-A homolog.J Cell Sci. 2024 Sep 15;137(18):jcs261895. doi: 10.1242/jcs.261895. Epub 2024 Sep 18. J Cell Sci. 2024. PMID: 39037215 Free PMC article.
-
Formation of artificial chromosomes in Caenorhabditis elegans and analyses of their segregation in mitosis, DNA sequence composition and holocentromere organization.Nucleic Acids Res. 2021 Sep 20;49(16):9174-9193. doi: 10.1093/nar/gkab690. Nucleic Acids Res. 2021. PMID: 34417622 Free PMC article.
References
-
- Van Hooser A.A., Ouspenski I.I., Gregson H.C., Starr D.A., Yen T.J., Goldberg M.L., Yokomori K., Earnshaw W.C., Sullivan K.F., Brinkley B.R.. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001; 114:3529–3542. - PubMed
-
- Harrington J.J., Van Bokkelen G., Mays R.W., Gustashaw K., Willard H.F.. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 1997; 15:345–355. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

