Cross-Species Metabolic Profiling of Floral Specialized Metabolism Facilitates Understanding of Evolutional Aspects of Metabolism Among Brassicaceae Species
- PMID: 33868339
- PMCID: PMC8045754
- DOI: 10.3389/fpls.2021.640141
Cross-Species Metabolic Profiling of Floral Specialized Metabolism Facilitates Understanding of Evolutional Aspects of Metabolism Among Brassicaceae Species
Abstract
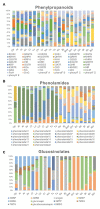
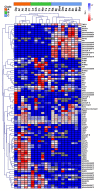
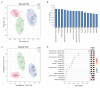
Plants produce a variety of floral specialized (secondary) metabolites with roles in several physiological functions, including light-protection, attraction of pollinators, and protection against herbivores. Pigments and volatiles synthesized in the petal have been focused on and characterized as major chemical factors influencing pollination. Recent advances in plant metabolomics have revealed that the major floral specialized metabolites found in land plant species are hydroxycinnamates, phenolamides, and flavonoids albeit these are present in various quantities and encompass diverse chemical structures in different species. Here, we analyzed numerous floral specialized metabolites in 20 different Brassicaceae genotypes encompassing both different species and in the case of crop species different cultivars including self-compatible (SC) and self-incompatible (SI) species by liquid chromatography-mass spectrometry (LC-MS). Of the 228 metabolites detected in flowers among 20 Brassicaceae species, 15 metabolite peaks including one phenylacyl-flavonoids and five phenolamides were detected and annotated as key metabolites to distinguish SC and SI plant species, respectively. Our results provide a family-wide metabolic framework and delineate signatures for compatible and incompatible genotypes thereby providing insight into evolutionary aspects of floral metabolism in Brassicaceae species.
Keywords: Brassicaceae; chemodiversity; compatible and incompatible species; cross-species comparison; flavonoids; floral specialized metabolite; plant metabolomics.
Copyright © 2021 Liu, Watanabe, Yasukawa, Kawamura, Aneklaphakij, Fernie and Tohge.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
High-performance liquid chromatography-mass spectrometry analysis of plant metabolites in brassicaceae.Methods Mol Biol. 2012;860:111-28. doi: 10.1007/978-1-61779-594-7_8. Methods Mol Biol. 2012. PMID: 22351174
-
Relaxed pollinator-mediated selection weakens floral integration in self-compatible taxa of Leavenworthia (Brassicaceae).Am J Bot. 2006 Jun;93(6):860-7. doi: 10.3732/ajb.93.6.860. Am J Bot. 2006. PMID: 21642148
-
Is floral structure a reliable indicator of breeding system in the Brassicaceae?PLoS One. 2017 Mar 21;12(3):e0174176. doi: 10.1371/journal.pone.0174176. eCollection 2017. PLoS One. 2017. PMID: 28323862 Free PMC article.
-
Brassicaceae flowers: diversity amid uniformity.J Exp Bot. 2019 May 9;70(10):2623-2635. doi: 10.1093/jxb/erz079. J Exp Bot. 2019. PMID: 30824938 Review.
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
Cited by
-
Environmental and Genetic Factors Involved in Plant Protection-Associated Secondary Metabolite Biosynthesis Pathways.Front Plant Sci. 2022 Apr 8;13:877304. doi: 10.3389/fpls.2022.877304. eCollection 2022. Front Plant Sci. 2022. PMID: 35463424 Free PMC article. Review.
References
-
- Ammar S., Abidi J., Luca S. V., Boumendjel M., Skalicka-Wozniak K., Bouaziz M. (2020). Untargeted metabolite profiling and phytochemical analysis based on RP-HPLC-DAD-QTOF-MS and MS/MS for discovering new bioactive compounds in Rumex algeriensis flowers and stems. Phytochem. Anal. 31, 616–635. 10.1002/pca.2928, PMID: - DOI - PubMed
-
- Anstey T. (1954). Self-incompatibility in green sprouting broccoli (Brassica oleracea L. Var. italica Plenck): I. Its occurrence and possible use in a breeding program. Can. J. Agric. Sci. 34, 59–64. 10.4141/agsci-1954-0009 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

