Dilated-DenseNet For Macromolecule Classification In Cryo-electron Tomography
- PMID: 33860285
- PMCID: PMC8046028
- DOI: 10.1007/978-3-030-57821-3_8
Dilated-DenseNet For Macromolecule Classification In Cryo-electron Tomography
Abstract
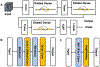
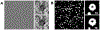
Cryo-electron tomography (cryo-ET) combined with subtomogram averaging (STA) is a unique technique in revealing macromolecule structures in their near-native state. However, due to the macromolecular structural heterogeneity, low signal-to-noise-ratio (SNR) and anisotropic resolution in the tomogram, macromolecule classification, a critical step of STA, remains a great challenge. In this paper, we propose a novel convolution neural network, named 3D-Dilated-DenseNet, to improve the performance of macromolecule classification in STA. The proposed 3D-Dilated-DenseNet is challenged by the synthetic dataset in the SHREC contest and the experimental dataset, and compared with the SHREC-CNN (the state-of-the-art CNN model in the SHREC contest) and the baseline 3D-DenseNet. The results showed that 3D-Dilated-DenseNet significantly outperformed 3D-DenseNet but 3D-DenseNet is well above SHREC-CNN. Moreover, in order to further demonstrate the validity of dilated convolution in the classification task, we visualized the feature map of 3D-Dilated-DenseNet and 3D-DenseNet. Dilated convolution extracts a much more representative feature map.
Keywords: Convolutional Neural Network; Cryo-electron Tomography; Object Classification; Subtomogram Averaging.
Figures
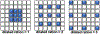
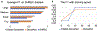

Similar articles
-
Macromolecules Structural Classification With a 3D Dilated Dense Network in Cryo-Electron Tomography.IEEE/ACM Trans Comput Biol Bioinform. 2022 Jan-Feb;19(1):209-219. doi: 10.1109/TCBB.2021.3065986. Epub 2022 Feb 3. IEEE/ACM Trans Comput Biol Bioinform. 2022. PMID: 33729943 Free PMC article.
-
VP-Detector: A 3D multi-scale dense convolutional neural network for macromolecule localization and classification in cryo-electron tomograms.Comput Methods Programs Biomed. 2022 Jun;221:106871. doi: 10.1016/j.cmpb.2022.106871. Epub 2022 May 11. Comput Methods Programs Biomed. 2022. PMID: 35584579
-
Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4Å.J Struct Biol. 2017 Sep;199(3):187-195. doi: 10.1016/j.jsb.2017.07.007. Epub 2017 Jul 22. J Struct Biol. 2017. PMID: 28743638 Free PMC article.
-
Subtomogram averaging from cryo-electron tomograms.Methods Cell Biol. 2019;152:217-259. doi: 10.1016/bs.mcb.2019.04.003. Epub 2019 May 15. Methods Cell Biol. 2019. PMID: 31326022 Review.
-
Cryo-Electron Tomography and Subtomogram Averaging.Methods Enzymol. 2016;579:329-67. doi: 10.1016/bs.mie.2016.04.014. Epub 2016 Jun 22. Methods Enzymol. 2016. PMID: 27572733 Review.
Cited by
-
FSCC: Few-Shot Learning for Macromolecule Classification Based on Contrastive Learning and Distribution Calibration in Cryo-Electron Tomography.Front Mol Biosci. 2022 Jul 5;9:931949. doi: 10.3389/fmolb.2022.931949. eCollection 2022. Front Mol Biosci. 2022. PMID: 35865006 Free PMC article.
-
Cryo-shift: reducing domain shift in cryo-electron subtomograms with unsupervised domain adaptation and randomization.Bioinformatics. 2022 Jan 27;38(4):977-984. doi: 10.1093/bioinformatics/btab794. Bioinformatics. 2022. PMID: 34897387 Free PMC article.
-
High-throughput cryo-ET structural pattern mining by unsupervised deep iterative subtomogram clustering.Proc Natl Acad Sci U S A. 2023 Apr 11;120(15):e2213149120. doi: 10.1073/pnas.2213149120. Epub 2023 Apr 7. Proc Natl Acad Sci U S A. 2023. PMID: 37027429 Free PMC article.
References
-
- Grünewald K, Medalia O, Gross A, Steven AC, Baumeister W: Prospects of electron cryotomography to visualize macromolecular complexes inside cellular compartments: implications of crowding. Biophysical chemistry 100(1-3) (2002) 577–591 - PubMed
-
- Han R, Wan X, Wang Z, Hao Y, Zhang J, Chen Y, Gao X, Liu Z, Ren F, Sun F, et al.: Autom: a novel automatic platform for electron tomography reconstruction. Journal of structural biology 199(3) (2017) 196–208 - PubMed
-
- Han R, Wang L, Liu Z, Sun F, Zhang F: A novel fully automatic scheme for fiducial marker-based alignment in electron tomography. Journal of structural biology 192(3) (2015) 403–417 - PubMed
-
- Wan W, Briggs J: Cryo-electron tomography and subtomogram averaging. In: Methods in enzymology. Volume 579. Elsevier; (2016) 329–367 - PubMed
-
- Ortiz JO, Förster F, Kürner J, Linaroudis AA, Baumeister W: Mapping 70s ribosomes in intact cells by cryoelectron tomography and pattern recognition. Journal of structural biology 156(2) (2006) 334–341 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
