Spatial Organization of Chromatin: Transcriptional Control of Adaptive Immune Cell Development
- PMID: 33854505
- PMCID: PMC8039525
- DOI: 10.3389/fimmu.2021.633825
Spatial Organization of Chromatin: Transcriptional Control of Adaptive Immune Cell Development
Abstract
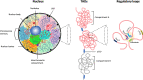
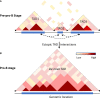
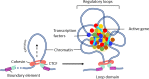
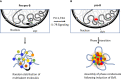
Higher-order spatial organization of the genome into chromatin compartments (permissive and repressive), self-associating domains (TADs), and regulatory loops provides structural integrity and offers diverse gene regulatory controls. In particular, chromatin regulatory loops, which bring enhancer and associated transcription factors in close spatial proximity to target gene promoters, play essential roles in regulating gene expression. The establishment and maintenance of such chromatin loops are predominantly mediated involving CTCF and the cohesin machinery. In recent years, significant progress has been made in revealing how loops are assembled and how they modulate patterns of gene expression. Here we will discuss the mechanistic principles that underpin the establishment of three-dimensional (3D) chromatin structure and how changes in chromatin structure relate to alterations in gene programs that establish immune cell fate.
Keywords: B and T cell development; chromatin organization; cis-regulatory interactions; gene regulatory networks; phase-separation.
Copyright © 2021 Pongubala and Murre.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Organizational principles of 3D genome architecture.Nat Rev Genet. 2018 Dec;19(12):789-800. doi: 10.1038/s41576-018-0060-8. Nat Rev Genet. 2018. PMID: 30367165 Free PMC article. Review.
-
Recent evidence that TADs and chromatin loops are dynamic structures.Nucleus. 2018 Jan 1;9(1):20-32. doi: 10.1080/19491034.2017.1389365. Epub 2017 Dec 14. Nucleus. 2018. PMID: 29077530 Free PMC article. Review.
-
Forces driving the three-dimensional folding of eukaryotic genomes.Mol Syst Biol. 2018 Jun 1;14(6):e8214. doi: 10.15252/msb.20188214. Mol Syst Biol. 2018. PMID: 29858282 Free PMC article. Review.
-
Myc Regulates Chromatin Decompaction and Nuclear Architecture during B Cell Activation.Mol Cell. 2017 Aug 17;67(4):566-578.e10. doi: 10.1016/j.molcel.2017.07.013. Epub 2017 Aug 10. Mol Cell. 2017. PMID: 28803781 Free PMC article.
-
Evolutionarily Conserved Principles Predict 3D Chromatin Organization.Mol Cell. 2017 Sep 7;67(5):837-852.e7. doi: 10.1016/j.molcel.2017.07.022. Epub 2017 Aug 17. Mol Cell. 2017. PMID: 28826674 Free PMC article.
Cited by
-
A regulatory network of microRNAs confers lineage commitment during early developmental trajectories of B and T lymphocytes.Proc Natl Acad Sci U S A. 2021 Nov 16;118(46):e2104297118. doi: 10.1073/pnas.2104297118. Proc Natl Acad Sci U S A. 2021. PMID: 34750254 Free PMC article.
-
Super-enhancers: drivers of cells' identities and cells' debacles.Epigenomics. 2024 Apr 8;16(9):681-700. doi: 10.2217/epi-2023-0409. Online ahead of print. Epigenomics. 2024. PMID: 38587919 Review.
-
Chromatin structure undergoes global and local reorganization during murine dendritic cell development and activation.Proc Natl Acad Sci U S A. 2022 Aug 23;119(34):e2207009119. doi: 10.1073/pnas.2207009119. Epub 2022 Aug 15. Proc Natl Acad Sci U S A. 2022. PMID: 35969760 Free PMC article.
-
Association of Single Nucleotide Polymorphisms in LEP, LEPR, and PPARG With Humoral Immune Response to Influenza Vaccine.Front Genet. 2021 Oct 22;12:725538. doi: 10.3389/fgene.2021.725538. eCollection 2021. Front Genet. 2021. PMID: 34745208 Free PMC article.
-
Single-cell detection of primary transcripts, their genomic loci and nuclear factors by 3D immuno-RNA/DNA FISH in T cells.Front Immunol. 2023 May 4;14:1156077. doi: 10.3389/fimmu.2023.1156077. eCollection 2023. Front Immunol. 2023. PMID: 37215121 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

